Difference between revisions of "Journal:Emerging and established trends to support secure health information exchange"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 100: | Line 100: | ||
* In the area of DNA sequencing and other -omics data formats, we have FASTQ file format<ref name="CockTheSanger10">{{cite journal |title=The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants |journal=Nucleic Acids Research |author=Cock, P.J.A.; Fields, C.J.; Goto, N. et al. |volume=38 |issue=6 |pages=1767–71 |year=2010 |doi=10.1093/nar/gkp1137 |pmid=20015970 |pmc=PMC2847217}}</ref>, which is used to store sequence information, and the standard flowgram format (SFF), which is used to encode sequence reads.<ref name="LeinonenTheSeq11">{{cite journal |title=The sequence read archive |journal=Nucleic Acids Research |author=Leinonen, R.; Sugawara, H.; Shumway, M. et al. |volume=39 |issue=DB1 |pages=D19–21 |year=2011 |doi=10.1093/nar/gkq1019 |pmid=21062823 |pmc=PMC3013647}}</ref> | * In the area of DNA sequencing and other -omics data formats, we have FASTQ file format<ref name="CockTheSanger10">{{cite journal |title=The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants |journal=Nucleic Acids Research |author=Cock, P.J.A.; Fields, C.J.; Goto, N. et al. |volume=38 |issue=6 |pages=1767–71 |year=2010 |doi=10.1093/nar/gkp1137 |pmid=20015970 |pmc=PMC2847217}}</ref>, which is used to store sequence information, and the standard flowgram format (SFF), which is used to encode sequence reads.<ref name="LeinonenTheSeq11">{{cite journal |title=The sequence read archive |journal=Nucleic Acids Research |author=Leinonen, R.; Sugawara, H.; Shumway, M. et al. |volume=39 |issue=DB1 |pages=D19–21 |year=2011 |doi=10.1093/nar/gkq1019 |pmid=21062823 |pmc=PMC3013647}}</ref> | ||
* The majority of the EHR systems adopt the HL7 standard clinical document architecture (CDA) as the interoperable data format.<ref name="DolinTheHL7_01">{{cite journal |title=The HL7 Clinical Document Architecture |journal=JAMIA |author=Dolin, R.H.; Alschuler, L.; Beebe, C. et al. |volume=8 |issue=6 |pages=552–69 |year=2001 |doi=10.1136/jamia.2001.0080552 |pmid=11687563 |pmc=PMC130066}}</ref> CDA is part of the HL7 Version 3 family, and it is based on a reference information model (RIM) that serves as a semantic model that consists of a set of structural components (e.g., classes with data types) and semantic relations that are used to represent clinical notes in the form of an extensible markup language document. | * The majority of the EHR systems adopt the HL7 standard clinical document architecture (CDA) as the interoperable data format.<ref name="DolinTheHL7_01">{{cite journal |title=The HL7 Clinical Document Architecture |journal=JAMIA |author=Dolin, R.H.; Alschuler, L.; Beebe, C. et al. |volume=8 |issue=6 |pages=552–69 |year=2001 |doi=10.1136/jamia.2001.0080552 |pmid=11687563 |pmc=PMC130066}}</ref> CDA is part of the HL7 Version 3 family, and it is based on a reference information model (RIM) that serves as a semantic model that consists of a set of structural components (e.g., classes with data types) and semantic relations that are used to represent clinical notes in the form of an extensible markup language document. | ||
The use of controlled vocabularies and terminologies allows for the unambiguous representation of important value sets such as "diagnosis" and "prescribed medicines."<ref name="Freitas">{{cite journal |title=Survey of current terminologies and ontologies in biology and medicine |journal=RECIIS |author=Freitas, F.; Schulz, S.; Moraes, E. |volume=3 |issue=1 |pages=239–49 |year=2009 |doi=10.3395/reciis.v3i1.239en}}</ref> The following are examples of such terminologies: | |||
* The [[International Statistical Classification of Diseases and Related Health Problems|International Classification of Diseases]] (ICD) provides a common language for reporting and monitoring diseases, used throughout the world to compare and share data in a consistent standard way between hospitals, regions, and countries and over periods of time. It is used to classify diseases and other problems for payment, management, and research, as recorded on many types of health records including medical records and death certificates. ICD-11 is the latest version of it, whereas ICD-10 (released in 1993) remains widely used. | |||
* SNOMED CT, already mentioned above, is the most comprehensive multilingual clinical healthcare terminology available. It is used in EHR systems to facilitate clinical documentation and reporting and to retrieve and analyze clinical data. SNOMED CT is both a coding scheme, identifying concepts and terms, and a multidimensional classification, enabling concepts to be related to each other, grouped, and analyzed according to different criteria. | |||
* Logical Observation Identifiers Names and Codes ([[LOINC]]) provides a set of universal identifiers for medical laboratory observations. LOINC provides codes for the observation names (e.g., eye color), not the observation finding (e.g., blue eyes). LOINC therefore provides codes for questions, and where needed, other vocabularies, such as SNOMED CT, provide codes for the answers. | |||
* The [[Unified Medical Language System]] (UMLS) is an important terminology resource, intended for use mainly by developers of health information systems. The UMLS “Metathesaurus” uses several different source vocabularies and seeks to reflect and preserve the meanings of concept names and relationships from these sources. It is therefore a valuable resource for the translation between the different source vocabularies. | |||
Application-level interfaces are also needed to support the communication and exchange of the standards-based encoded information. The role of HL7 is principal on this front: HL7's name comes from “Level Seven,” which, according to the Open Systems Interconnection (OSI) model that standardizes communication functionality in IT, corresponds to application layer. From its establishment in the late 1980s, HL7 was therefore focused on exchanging information within hospitals. The focus remains almost the same today, but HL7 has progressed from different paradigms over the years, in order to describe the structure, semantics, and management of the exchanged information. The development of HL7 Version 3 (HL7v3) started around 1995 in order to introduce more consistency between the implementations of Version 2 following an object-oriented development methodology. The most recent proposal by HL7 is [[Fast Healthcare Interoperability Resources]] (FHIR), which leverages web technologies to overcome the complexity of HL7v3.<ref name="BenderHL713">{{cite journal |title=HL7 FHIR: An Agile and RESTful approach to healthcare information exchange |journal=Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems |author=Bender, D.; Sartipi, K. |pages=326–31 |year=2013 |doi=10.1109/CBMS.2013.6627810}}</ref> | |||
On the other hand, the IHE initiative has defined a number of “integration profiles,” which are detailed specifications for communication among systems to address key clinical use cases, all based on established standards. IHE profiles organize and leverage the integration capabilities that can be achieved by coordinated implementation of communication standards, such as DICOM, HL7, W3C, and other security standards.<ref name="KondylakisUsing19">{{cite journal |title=Using XDS and FHIR to Support Mobile Access to EHR Information Through Personal Health Apps |journal=Proceedings of the 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems |author=Kondylakis, H.; Petrakis, Y.; Leivadoros, S. et al. |pages=241–4 |year=2019 |doi=10.1109/CBMS.2019.00058}}</ref> Some of the IHE integration profiles that might be interesting in the context of interfacing health information systems and sharing of clinical information include: | |||
* Cross-Enterprise Document Sharing (XDS): Allows EHR documents to be shared and discovered among healthcare enterprises, physician offices, clinics, acute care in-patient facilities, and PHRs. | |||
* Patient Demographics Query (PDQ): Enables applications to query by patient demographics (e.g., name) for patient identity from a central patient information server. | |||
* Patient Identifier Cross Referencing (PIX): Allows applications to query for patient identity cross-references between hospitals, sites, HIE networks, etc. | |||
* PDQ HL7 v3 (PDQv3): Extends the PDQ profile leveraging HL7 Version 3. | |||
* PIX: Extends the Patient Identifier Cross-Reference profile leveraging HL7 Version 3. | |||
* Cross-Community Access (XCA): Allows the query and retrieval of patients' EHRs held by other communities. | |||
* Cross-Enterprise Document Reliable Interchange (XDR): Exchanges health documents between health enterprises using a web service-based point-to-point push network communication. | |||
There are two main architectural approaches for the implementation of an information sharing platform: a centralized approach and a federated approach.<ref name="EckmanVarieties07">{{cite journal |title=Varieties of interoperability in the transformation of the health-care information infrastructure |journal=IBM Systems Journal |author=Eckman, B.A.; Bennett, C.A.; Kaufman, J.H. et al. |volume=46 |issue=1 |pages=19–41 |year=2007 |doi=10.1147/sj.461.0019}}</ref> In the centralized approach, a central data warehouse and accompanying services act as middlemen for the exchange of information and a single source of patient data that are shared among the participating organizations. On the other hand, in the federated architecture, a central infrastructure is also in place, but in this case, it merely acts as a facilitator for locating the data sources. An example of this case would be a common registry that stores only the links to the original patient records, medical images, and other types of data while the linked data are not transferred outside their primary premises unless explicitly requested by any interested client system. In addition to these opposite approaches for designing a distributed information sharing platform, there are also various hybrid options, such as using messaging with “publish-subscribe” communication that can be introduced to complement either the centralized or federated architectures. | |||
There are advantages and disadvantages in all of the abovementioned deployment options. For example, in the federated approach, there are more strong concerns about the privacy, security, and availability of the data shared and their original sources. (26) The operation of a mission-critical [[radiology information system]] (RIS) in a hospital can be severely affected if multiple peers request DICOM images from its PACS, and this poses an additional burden and cost for the acquisition and management of adequate infrastructure in the source organization. Instead, a centralized strategy allows for easy access to the whole information shared but also leads to a concentration of the costs for maintaining the infrastructure needed and can be problematic at the operation level (a “single point of failure”). Furthermore, there are more costs on integrating the different data sets under a common “schema,” resolving conflicts or even supporting the timely update of the persisted information when a source system acquires new or modified data. | |||
==Emerging supportive technologies: Blockchain== | |||
Revision as of 17:02, 8 June 2021
| Full article title | Emerging and established trends to support secure health information exchange |
|---|---|
| Journal | Frontiers in Digital Health |
| Author(s) | Spanakis, Emmanouil G.; Sfakianakis, Stelios; Bonomi, Silvia; Ciccotelli, Claudio; Magalini, Sabina; Sakkalis, Vangelis |
| Author affiliation(s) | Foundation for Research and Technology, Sapienza Università di Roma, Fondazione Policlinico Universitario Agostino Gemelli |
| Primary contact | Email: spanakis at ics dot forth dot gr |
| Editors | Pattichis, Constantinos S. |
| Year published | 2021 |
| Volume and issue | 3 |
| Article # | 636082 |
| DOI | 10.3389/fdgth.2021.636082 |
| ISSN | 2673-253X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fdgth.2021.636082/full |
| Download | https://www.frontiersin.org/articles/10.3389/fdgth.2021.636082/pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
This work aims to provide information, guidelines, established practices and standards, and an extensive evaluation on new and promising technologies for the implementation of a secure information sharing platform for health-related data. We focus strictly on the technical aspects and specifically on the sharing of health information, studying innovative techniques for secure information sharing within the healthcare domain, and we describe our solution and evaluate the use of blockchain methodologically for integrating within our implementation. To do so, we analyze health information sharing within the concept of the PANACEA project, which facilitates the design, implementation, and deployment of a relevant platform. The research presented in this paper provides evidence and argumentation toward advanced and novel implementation strategies for a state-of-the-art information sharing environment; a description of high-level requirements for the national and cross-border transfer of data between different healthcare organizations; technologies to support the secure interconnectivity and trust between information technology (IT) systems participating in a data-sharing “community”; standards, guidelines, and interoperability specifications for implementing a common understanding and integration in the sharing of clinical information; and the use of cloud computing and prospectively more advanced technologies such as blockchain. The technologies described and the possible implementation approaches are presented in the design of an innovative secure information sharing platform in the healthcare domain.
Introduction
Information technology (IT) has long been identified as a cornerstone for efficient, cost-saving, timely, and reliable healthcare delivery.[1][2] The availability of healthcare information and patient records in digital form facilitates the persistence and posterity of valuable information and greatly supports the decision-making process, and even the extraction of new knowledge at both the individual and population levels. In our previous work, we have emphasized the current state of the art about cybersecurity in the healthcare domain, with emphasis on current threats and methodologies.[3] Paraphrasing the famous words of John Donne, “no IT system is an island, entire of itself.” Today, in a highly connected world where geographic boundaries have been largely eliminated and people can freely move between cities, states, countries, or continents, the requirement for two different information management systems to exchange a person's clinical data or medical history becomes vital and persistent. Sharing health information (e.g., via health information exchange [HIE]) through electronic means greatly improves the cost, quality, and patient experience of healthcare delivery.
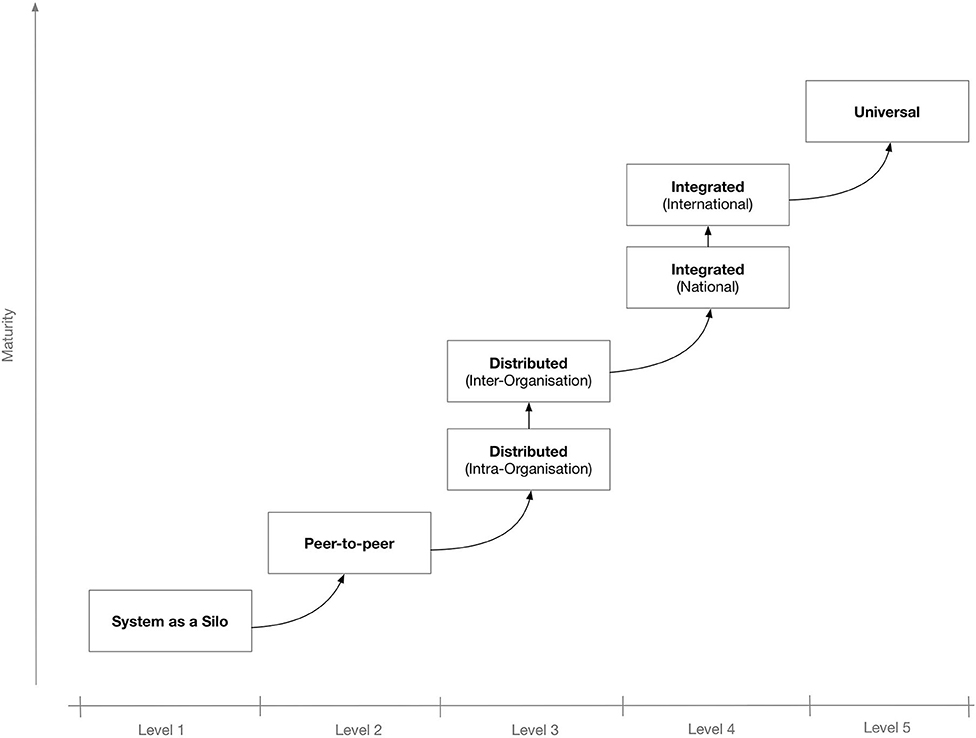
To better secure the IT system's potential for interconnectivity and cooperation with other systems, the use of interoperable technologies and standards is needed. Depending on the extent and scope of the envisaged shared information spaces, there may be different levels of interoperability. Figure 1 shows a proposed “maturity” model for interoperability in eHealth.[4] The model consists of five levels that, incrementally, describe a more mature version of an interoperable infrastructure, starting from Level 1 for non-connected eHealth applications; Level 2 where a single eHealth application is directly linked to another application for simple data exchange[5]; Level 3 for distributed systems that agree on protocols used, data formats, message exchange patterns, etc.[6]; Level 4, where eHealth applications from different suppliers that serve a common goal are linked but the applications do not need to have common objectives[7]; and finally, at the “universal” Level 5, where diverse eHealth applications connect to an open, interoperable infrastructure possibly spanning multiple countries.[8][9]
|
Interoperability and data sharing in the healthcare domain is additionally challenging due to the multiplicity of the stakeholders, that is, the entities that operate (or are involved in any way) in this domain and which will be affected by any “disruption” or reform of the system. Some of the most important stakeholders or actors are therefore the following:
- the patients who are actually treated or, in general, are the recipients of the health services;
- the medical professionals (physicians and medical personnel) to provide the medical care;
- the healthcare organizations (HCOs) (healthcare providers) as represented by their board of directors, who actually administer the health delivery from a business perspective;
- the insurance companies that provide health coverage plans;
- the pharmaceutical companies that produce and market medications to be prescribed by physicians for the treatment of patients; and
- the governments and other regulatory parties who control, coordinate, and set the rules, rights, and obligations of any involved party.
All these actors could have an influence in the design of a data sharing system and can also set important—and conflicting in some cases—requirements. For example:
- Patients would like to have their medical record shared, but only after their approval and only with specific authorized personnel in specific circumstances.
- An HCO can be extremely cautious about sharing the data of their patients with another organization because they are concerned by the security and availability of their systems.
- Governments of E.U. member states can impose strict laws about the transfer of their citizens in cross-border healthcare treatment scenarios.
- Medical professionals require fast and effortless access to a patient's medical history in emergency situations, which cannot be the case if time-consuming authorization processes are the norm.
Given these and similar requirements, and even though the objective is to design a technical solution for the sharing of clinical data, it is imperative that all these constraints and requirements are considered and addressed in a satisfying manner.
From a strictly technical point of view, the sharing platform may need to be interoperable with a large number and diverse set of IT systems, each with their own protocols, data formats, etc. Some of the most important systems that manage patient-related data, and could be used as data sources for information sharing, include electronic health records (EHRs), personal health records (PHRs), laboratory information systems (LISs), and picture archiving and communication systems (PACS). EHRs are patient-centered systems that store and manage clinical information, such as a patient's medical history, diagnoses, medications, immunization dates, allergies, radiology images, and lab and test results. They are managed by authorized personnel, usually in the context of a single HCO, although they can span beyond that. PHRs are electronic applications that are used by people managing their own health information in a private and confidential environment. They are simpler systems than EHRs, and in some cases, they can be connected (temporarily or otherwise) to more enterprise-level HCO systems (e.g., EHRs or other hospital information systems). An LIS is used inside hospitals and clinics to record, manage, and store data for clinical laboratories in a patient-centric way (e.g., sending laboratory test orders to lab instruments, tracking those orders, and then recording the results in a searchable database). And PACS are systems used in a clinical setting for the storage and convenient access to medical images from multiple modalities (source machine types). Digital Imaging and Communications in Medicine (DICOM) is the standard format and suite of protocols for the storage and transfer of images from PACS services. Of course, in a health ecosystem there may be additional systems, for example, for the management of insurance claims and for clinidal diagnoses (e.g., clinical decision support systems [CDSSs]).
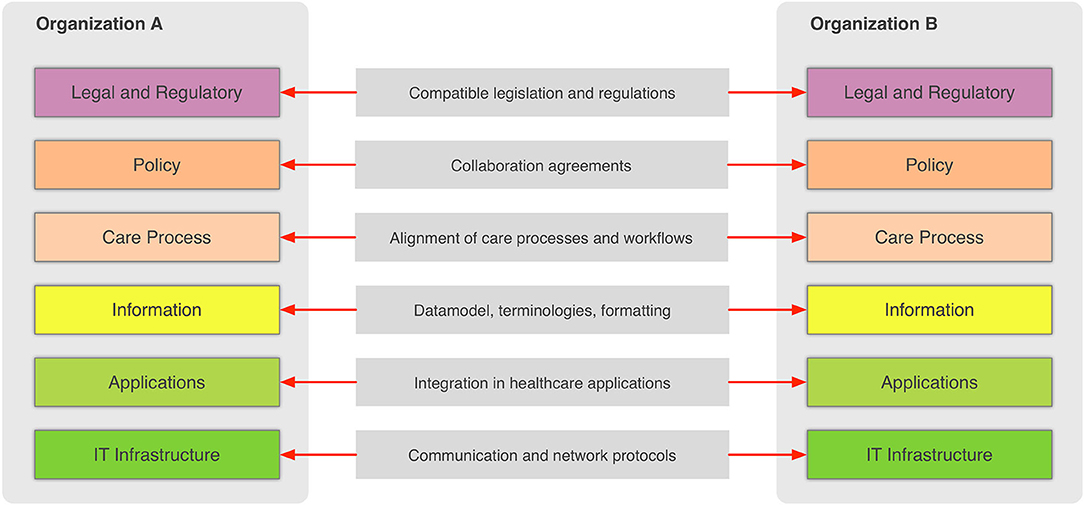
In the general context, information sharing involves more than one party (healthcare providers, organizations, etc.,) that need to cooperate and agree on the way the exchange of information happens, and what the rules and policies are that govern it. Interoperability involves many different aspects, such as legislation and guidelines, contracts, and agreements between exchanging parties, governance and maintenance, shareable workflows, standardized data elements, semantic and syntactic choices, applications, technical infrastructure, and safety and privacy issues. The Refined eHealth European Interoperability Framework (EIF) is a set of recommendations that specify the standards, protocols, procedures, and policies that when deployed can improve the interoperability of eHealth applications within the E.U. and across its member states by providing specific recommendations for all these aspects.[10] Figure 2 depicts how these aspects can be represented in interoperability “levels” that permit two different organizations to communicate.
|
This framework provides a great overview of the needed “glue” so that two or more healthcare environments can collaborate and serve as a common, multilevel, and multi-perspective model on the interoperability requirements. The six different levels of the (refined) EIF are:
- Legal and regulatory: This level represents the legislation and regulatory guidelines that define the boundaries for interoperability not only across borders, but also within a country or region.
- Policy: This level represents the contracts and agreements between the sharing organizations so that trust is established and responsibilities are assigned.
- Care process: This level represents the shared workflows that define how the integrated care is delivered and how these workflows are managed.
- Information: This level defines the data models, the concepts and their values, the terminologies, and controlled vocabularies that cater for the common understanding of the exchanged electronic messages.
- Applications: This level defines how the data are extracted from and imported to the healthcare information systems and how the transport of the data takes place using health-specific technologies and standards.
- IT infrastructure: This level represents the lowest level and corresponds to general-purpose communication and network protocols.
This generic EIF is highly relevant for the use cases of the health information sharing since sharing is greatly facilitated between interoperable systems and organizations. Here, we focus on the sharing of health-related information across HCOs and even across countries and continents. This is an important use case to improve the secure and efficient delivery of health care across Europe.[11][12] The importance of cross-border health care in Europe has been recognized since 2011's Directive 2011/24/EU, which established patients' rights to access safe and high-quality healthcare, including across national borders within the E.U., and their right to be reimbursed for such healthcare.[13] As can be deduced by considering the Refined eHealth EIF (Figure 2), the cross-border sharing of clinical information is a complex scenario due to the fact that data need to be transferred between different countries and therefore, requires overcoming barriers such as establishing a common trust framework, uniquely identifying citizens, and translating between different schemas and terminologies. This paper presents current approaches to address most of these issues in the European context while presenting and evaluating emerging technologies such as blockchain that have been in the limelight recently. Our objective is to take advantage of both well-established and novel technologies that complement each other in order to design an architecture for the secure exchange of clinical information in the European context.
Materials and methods
Current status on standards-based health data exchange
In the healthcare industry, large standards developing organizations have defined numerous standards, data formats, terminologies, and more in order to support the design and building of interoperable IT systems. Perhaps the most well-known and most important standards are the ones introduced by Health Level 7 (HL7) and SNOMED CT, which can be used as a foundation for the development of data exchange standards among eHealth systems.[14] Two more standards organizations are Integrating the Health Enterprise (IHE), which focuses primarily on integration and interoperability, and the Clinical Data Interchange Standards Consortium (CDISC). CDISC produced the Operational Data Model (ODM) to “facilitate the regulatory-compliant acquisition, archive, and interchange of metadata and data for clinical research studies.”[15] ODM is an XML-based format that provides a number of constructs for modeling electronic case report forms (CRFs) and can also be used in sending forms data from a clinical trial system to an EHR system. In the area of medical devices, the Continua Health Alliance, a non-profit, open-industry coalition of healthcare and technology companies working to establish a system of interoperable personal health solutions, develops an ecosystem of connected technologies, devices, and services that will enable the more efficient exchange of fitness, health, and wellness information.[16] Among its proposed standards, Continua proposes specifications and standards such as Bluetooth, USB, ISO/IEEE 11073 (for medical devices), and HL7 to enable patients to use home-based devices to monitor their weight, blood pressure, and glucose and blood oxygen levels, as well as to share these data with their healthcare professionals.
The exchanged information can be in multiple data formats based on the type of data, device category, etc.[17] Some of the most common formats based on the health applications using them are the following:
- For medical imaging, the use of DICOM is almost universal and defines not only the content (DICOM file format) but also communication protocols for the exchange of medical images.[18]
- In the area of DNA sequencing and other -omics data formats, we have FASTQ file format[19], which is used to store sequence information, and the standard flowgram format (SFF), which is used to encode sequence reads.[20]
- The majority of the EHR systems adopt the HL7 standard clinical document architecture (CDA) as the interoperable data format.[21] CDA is part of the HL7 Version 3 family, and it is based on a reference information model (RIM) that serves as a semantic model that consists of a set of structural components (e.g., classes with data types) and semantic relations that are used to represent clinical notes in the form of an extensible markup language document.
The use of controlled vocabularies and terminologies allows for the unambiguous representation of important value sets such as "diagnosis" and "prescribed medicines."[22] The following are examples of such terminologies:
- The International Classification of Diseases (ICD) provides a common language for reporting and monitoring diseases, used throughout the world to compare and share data in a consistent standard way between hospitals, regions, and countries and over periods of time. It is used to classify diseases and other problems for payment, management, and research, as recorded on many types of health records including medical records and death certificates. ICD-11 is the latest version of it, whereas ICD-10 (released in 1993) remains widely used.
- SNOMED CT, already mentioned above, is the most comprehensive multilingual clinical healthcare terminology available. It is used in EHR systems to facilitate clinical documentation and reporting and to retrieve and analyze clinical data. SNOMED CT is both a coding scheme, identifying concepts and terms, and a multidimensional classification, enabling concepts to be related to each other, grouped, and analyzed according to different criteria.
- Logical Observation Identifiers Names and Codes (LOINC) provides a set of universal identifiers for medical laboratory observations. LOINC provides codes for the observation names (e.g., eye color), not the observation finding (e.g., blue eyes). LOINC therefore provides codes for questions, and where needed, other vocabularies, such as SNOMED CT, provide codes for the answers.
- The Unified Medical Language System (UMLS) is an important terminology resource, intended for use mainly by developers of health information systems. The UMLS “Metathesaurus” uses several different source vocabularies and seeks to reflect and preserve the meanings of concept names and relationships from these sources. It is therefore a valuable resource for the translation between the different source vocabularies.
Application-level interfaces are also needed to support the communication and exchange of the standards-based encoded information. The role of HL7 is principal on this front: HL7's name comes from “Level Seven,” which, according to the Open Systems Interconnection (OSI) model that standardizes communication functionality in IT, corresponds to application layer. From its establishment in the late 1980s, HL7 was therefore focused on exchanging information within hospitals. The focus remains almost the same today, but HL7 has progressed from different paradigms over the years, in order to describe the structure, semantics, and management of the exchanged information. The development of HL7 Version 3 (HL7v3) started around 1995 in order to introduce more consistency between the implementations of Version 2 following an object-oriented development methodology. The most recent proposal by HL7 is Fast Healthcare Interoperability Resources (FHIR), which leverages web technologies to overcome the complexity of HL7v3.[23]
On the other hand, the IHE initiative has defined a number of “integration profiles,” which are detailed specifications for communication among systems to address key clinical use cases, all based on established standards. IHE profiles organize and leverage the integration capabilities that can be achieved by coordinated implementation of communication standards, such as DICOM, HL7, W3C, and other security standards.[24] Some of the IHE integration profiles that might be interesting in the context of interfacing health information systems and sharing of clinical information include:
- Cross-Enterprise Document Sharing (XDS): Allows EHR documents to be shared and discovered among healthcare enterprises, physician offices, clinics, acute care in-patient facilities, and PHRs.
- Patient Demographics Query (PDQ): Enables applications to query by patient demographics (e.g., name) for patient identity from a central patient information server.
- Patient Identifier Cross Referencing (PIX): Allows applications to query for patient identity cross-references between hospitals, sites, HIE networks, etc.
- PDQ HL7 v3 (PDQv3): Extends the PDQ profile leveraging HL7 Version 3.
- PIX: Extends the Patient Identifier Cross-Reference profile leveraging HL7 Version 3.
- Cross-Community Access (XCA): Allows the query and retrieval of patients' EHRs held by other communities.
- Cross-Enterprise Document Reliable Interchange (XDR): Exchanges health documents between health enterprises using a web service-based point-to-point push network communication.
There are two main architectural approaches for the implementation of an information sharing platform: a centralized approach and a federated approach.[25] In the centralized approach, a central data warehouse and accompanying services act as middlemen for the exchange of information and a single source of patient data that are shared among the participating organizations. On the other hand, in the federated architecture, a central infrastructure is also in place, but in this case, it merely acts as a facilitator for locating the data sources. An example of this case would be a common registry that stores only the links to the original patient records, medical images, and other types of data while the linked data are not transferred outside their primary premises unless explicitly requested by any interested client system. In addition to these opposite approaches for designing a distributed information sharing platform, there are also various hybrid options, such as using messaging with “publish-subscribe” communication that can be introduced to complement either the centralized or federated architectures.
There are advantages and disadvantages in all of the abovementioned deployment options. For example, in the federated approach, there are more strong concerns about the privacy, security, and availability of the data shared and their original sources. (26) The operation of a mission-critical radiology information system (RIS) in a hospital can be severely affected if multiple peers request DICOM images from its PACS, and this poses an additional burden and cost for the acquisition and management of adequate infrastructure in the source organization. Instead, a centralized strategy allows for easy access to the whole information shared but also leads to a concentration of the costs for maintaining the infrastructure needed and can be problematic at the operation level (a “single point of failure”). Furthermore, there are more costs on integrating the different data sets under a common “schema,” resolving conflicts or even supporting the timely update of the persisted information when a source system acquires new or modified data.
Emerging supportive technologies: Blockchain
References
- ↑ U.S. Congress, Office of Technology Assessment (September 1995). "Bringing Health Care Online: The Role of Information Technologies" (PDF). U.S. Government Printing Office. https://ota.fas.org/reports/9507.pdf.
- ↑ Kolodner, R.M.; Cohn, S.P.; Freidman, C.P. (2008). "Health Information Technology: Strategic Initiatives, Real Progress". Health Affairs 27 (Supp. 1): w391–5. doi:10.1377/hlthaff.27.5.w391.
- ↑ Spanakis, E.G.; Bonomi, S.; Sfakianakis, S. (2020). "Cyber-attacks and threats for healthcare - A multi-layer thread analysis". Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society: 5705–08. doi:10.1109/EMBC44109.2020.9176698. PMID 33019270.
- ↑ 4.0 4.1 Van Velsen, L.; Hermens, H.; d'Hollosy, O.-N. (2016). "A maturity model for interoperability in eHealth". Proceedings of the IEEE 18th International Conference on e-Health Networking, Applications and Services: 1–6. doi:10.1109/HealthCom.2016.7749533.
- ↑ Chronaki, C.E.; Chiarugi, F.; Mavrogiannaki, E. et al. (2003). "An eHealth platform for instant interaction among health professionals". Proceedings of the Computers in Cardiology 2003: 101–4. doi:10.1109/CIC.2003.1291100.
- ↑ Spanakis, M; Lelis, P.; Chiarugi, F. et al. (2005). "R&D Challenges in Developing an Ambient Intelligence eHealth Platform". IFMBE Proceedings 11 (1): 1727–983. http://139.91.210.27/CBML/PROCEEDINGS/2005_EMBEC/Embec%202005/Abstracts/Abstract791.html.
- ↑ Tsiknakis, M.; Spanakis, M. (2010). "Adoption of innovative eHealth services in prehospital emergency management: A case study". Proceedings of the 10th IEEE International Conference on Information Technology and Applications in Biomedicine: 1–5. doi:10.1109/ITAB.2010.5687752.
- ↑ Spanakis, E.G.; Psaraki, M.; Sakkalis, V. (2018). "Congestive Heart Failure Risk Assessment Monitoring through Internet of things and mobile Personal Health Systems". Proceedings of the 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society: 2925-28. doi:10.1109/EMBC.2018.8513024. PMID 30441013.
- ↑ Spanakis, M.; Sfakianakas, S.; Sakkalis, V. et al. (2019). "PharmActa: Empowering Patients to Avoid Clinical Significant Drug⁻Herb Interactions". Medicines 6 (1): 26. doi:10.3390/medicines6010026. PMC PMC6473432. PMID 30781500. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6473432.
- ↑ "eHealth Network: Refined eHealth European Interoperability Framework" (PDF). European Union. 23 November 2015. pp. 24. https://ec.europa.eu/health/sites/default/files/ehealth/docs/ev_20151123_co03_en.pdf.
- ↑ Katehakis, D.G.; Pangalos, G.; Prentza, A. (2017). "Security Improvements for Better and Safer Cross-Border ePrescription and Patient Summary Services". International Journal of Reliable and Quality E-Healthcare 6 (1): 2. doi:10.4018/IJRQEH.2017010102.
- ↑ Tinholt, D.; Carrara, W.; Tol, T. et al. (18 June 2013). "Final Report - Study on Analysis of the Needs for Cross-Border Services and Assessment of the Organisational, Legal, Technical and Semantic Barriers (SMART 2011/0074)". European Commission. doi:10.2759/10003. https://ec.europa.eu/digital-single-market/en/news/final-report-study-analysis-needs-cross-border-services-and-assessment-organisational-legal.
- ↑ Peeters, M. (2012). "Free movement of patients: Directive 2011/24 on the application of patients' rights in cross-border healthcare". European Journal of Health Law 19 (1): 29–60. doi:10.1163/157180912x615158. PMID 22428388.
- ↑ Benson, T. (2012). Principles of Health Interoperability HL7 and SNOMED (2nd ed.). Springer. doi:10.1007/978-1-4471-2801-4. ISBN 9781447128014.
- ↑ Kuchinke, W.; Alerts, J.; Semler, S.C. et al. (2009). "CDISC standard-based electronic archiving of clinical trials". Methods of Information in Medicine 48 (5): 408–13. doi:10.3414/ME9236. PMID 19621114.
- ↑ Carroll, R.; Cnossen, R.; Schnell, M. et al. (2007). "Continua: An Interoperable Personal Healthcare Ecosystem". IEEE Pervasive Computing 6 (4): 90–94. doi:10.1109/MPRV.2007.72.
- ↑ Marias, K.; Sakkalis, A.; Roniotis, C. et al. (2009). "Clinically Oriented Translational Cancer Multilevel Modeling: The ContraCancrum Project". Proceedings of the 2009 World Congress on Medical Physics and Biomedical Engineering: 2121–27. doi:10.1007/978-3-642-03882-2_564.
- ↑ Larobina, M.; Murino, L. (2014). "Medical Image File Formats". Journal of Digital Imaging 27: 200–06. doi:10.1007/s10278-013-9657-9.
- ↑ Cock, P.J.A.; Fields, C.J.; Goto, N. et al. (2010). "The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants". Nucleic Acids Research 38 (6): 1767–71. doi:10.1093/nar/gkp1137. PMC PMC2847217. PMID 20015970. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2847217.
- ↑ Leinonen, R.; Sugawara, H.; Shumway, M. et al. (2011). "The sequence read archive". Nucleic Acids Research 39 (DB1): D19–21. doi:10.1093/nar/gkq1019. PMC PMC3013647. PMID 21062823. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3013647.
- ↑ Dolin, R.H.; Alschuler, L.; Beebe, C. et al. (2001). "The HL7 Clinical Document Architecture". JAMIA 8 (6): 552–69. doi:10.1136/jamia.2001.0080552. PMC PMC130066. PMID 11687563. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC130066.
- ↑ Freitas, F.; Schulz, S.; Moraes, E. (2009). "Survey of current terminologies and ontologies in biology and medicine". RECIIS 3 (1): 239–49. doi:10.3395/reciis.v3i1.239en.
- ↑ Bender, D.; Sartipi, K. (2013). "HL7 FHIR: An Agile and RESTful approach to healthcare information exchange". Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems: 326–31. doi:10.1109/CBMS.2013.6627810.
- ↑ Kondylakis, H.; Petrakis, Y.; Leivadoros, S. et al. (2019). "Using XDS and FHIR to Support Mobile Access to EHR Information Through Personal Health Apps". Proceedings of the 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems: 241–4. doi:10.1109/CBMS.2019.00058.
- ↑ Eckman, B.A.; Bennett, C.A.; Kaufman, J.H. et al. (2007). "Varieties of interoperability in the transformation of the health-care information infrastructure". IBM Systems Journal 46 (1): 19–41. doi:10.1147/sj.461.0019.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, though grammar and word usage was substantially updated for improved readability. In some cases important information was missing from the references, and that information was added.