Difference between revisions of "Journal:Kadi4Mat: A research data infrastructure for materials science"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 59: | Line 59: | ||
The so-called workflows are of particular importance in the ELN component. A "workflow" is a generic concept that describes a well-defined sequence of sequential or parallel steps, which are processed as automatically as possible. This can include the execution of an analysis tool or the control and data retrieval of an experimental device. To accommodate such heterogeneity, concrete steps must be implemented as flexibly as possible, since they are highly user- and application-specific. In Figure 1, the types of tools shown in the second layer are used as part of the workflows, so as to implement the actual functionality of the various steps. These can be roughly divided into analysis, visualization, transformation, and transportation tasks. In order to keep the application of these tools as generic as possible, a combination of provided and user-defined tools is accessed. From a user’s perspective, it must be possible to provide such tools in an easy manner, while the execution of each tool must take place in a secure and functional environment. This is especially true for existing tools—e.g., a simple MATLAB<ref name="MATLAB">{{cite web |url=https://www.mathworks.com/products/matlab.html |title=MATLAB |publisher=MathWorks |accessdate=19 January 2021}}</ref> script—which require certain dependencies to be executed and must be equipped with a suitable interface to be used within a workflow. Depending on their functionality, the tools must in turn access various technical infrastructures. In addition to the use of the repository and computing infrastructure, direct access to devices is also important for more complex data analyses. The automation of a typical workflow of experimenters is only fully possible if data and metadata, created by devices, can be captured. However, such an integration is not trivial due to a heterogeneous device landscape, as well as proprietary data formats and interfaces.<ref name="HawkerLab07">{{cite journal |title=Laboratory Automation: Total and Subtotal |journal=Clinics in Laboratory Medicine |author=Hawker, C.D. |volume=27 |issue=4 |pages=749–70 |year=2007 |doi=10.1016/j.cll.2007.07.010}}</ref><ref name="PotthoffProc19">{{cite journal |title=Procedures for systematic capture and management of analytical data in academia |journal=Analytica Chimica Acta: X |author=Potthoff, J.; Tremouilhac, P.; Hodapp, P. et al. |volume=1 |at=100007 |year=2019 |doi=10.1016/j.acax.2019.100007}}</ref> In Kadi4Mat, it should also be possible to use individual tools separately, where appropriate, i.e., outside a workflow. For example, when using the web-based interface, a visualization tool for a custom data format may be used to generate a preview of a datum that can be directly displayed in a web browser. | The so-called workflows are of particular importance in the ELN component. A "workflow" is a generic concept that describes a well-defined sequence of sequential or parallel steps, which are processed as automatically as possible. This can include the execution of an analysis tool or the control and data retrieval of an experimental device. To accommodate such heterogeneity, concrete steps must be implemented as flexibly as possible, since they are highly user- and application-specific. In Figure 1, the types of tools shown in the second layer are used as part of the workflows, so as to implement the actual functionality of the various steps. These can be roughly divided into analysis, visualization, transformation, and transportation tasks. In order to keep the application of these tools as generic as possible, a combination of provided and user-defined tools is accessed. From a user’s perspective, it must be possible to provide such tools in an easy manner, while the execution of each tool must take place in a secure and functional environment. This is especially true for existing tools—e.g., a simple MATLAB<ref name="MATLAB">{{cite web |url=https://www.mathworks.com/products/matlab.html |title=MATLAB |publisher=MathWorks |accessdate=19 January 2021}}</ref> script—which require certain dependencies to be executed and must be equipped with a suitable interface to be used within a workflow. Depending on their functionality, the tools must in turn access various technical infrastructures. In addition to the use of the repository and computing infrastructure, direct access to devices is also important for more complex data analyses. The automation of a typical workflow of experimenters is only fully possible if data and metadata, created by devices, can be captured. However, such an integration is not trivial due to a heterogeneous device landscape, as well as proprietary data formats and interfaces.<ref name="HawkerLab07">{{cite journal |title=Laboratory Automation: Total and Subtotal |journal=Clinics in Laboratory Medicine |author=Hawker, C.D. |volume=27 |issue=4 |pages=749–70 |year=2007 |doi=10.1016/j.cll.2007.07.010}}</ref><ref name="PotthoffProc19">{{cite journal |title=Procedures for systematic capture and management of analytical data in academia |journal=Analytica Chimica Acta: X |author=Potthoff, J.; Tremouilhac, P.; Hodapp, P. et al. |volume=1 |at=100007 |year=2019 |doi=10.1016/j.acax.2019.100007}}</ref> In Kadi4Mat, it should also be possible to use individual tools separately, where appropriate, i.e., outside a workflow. For example, when using the web-based interface, a visualization tool for a custom data format may be used to generate a preview of a datum that can be directly displayed in a web browser. | ||
In Figure 2, the current concept for the integration of workflows in Kadi4Mat is shown. Different steps of a workflow can be defined with a graphical node editor. Either a web-based or a desktop-based version of such an editor can be used, the latter running as an ordinary application on a local workstation. With the help of such an editor, the different steps or tools to be executed are defined, linked, and, most importantly, parameterized. The execution of a workflow can be started via an external component called "Process Manager." This component in turn manages several process engines, which take care of executing the workflows. The process engines potentially differ in their implementation and functionality. A simple process engine, for example, could be limited to a sequential execution order of the different tasks, while another one could execute independent tasks in parallel. All engines process the required steps based on the [[information]] stored in the workflow. With appropriate transport tools, the data and metadata required for each step, as well as the resulting output, can be exported or imported from Kadi4Mat using the existing interfaces of the research data infrastructure. With similar tools, the use of other external data sources becomes possible, and with it the possibility to handle large amounts of data via suitable exchange protocols. The use of locally stored data is also possible when running a workflow on a local workstation. | |||
[[File:Fig2 Brandt DataSciJourn21 20-1.png|800px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="800px" | |||
|- | |||
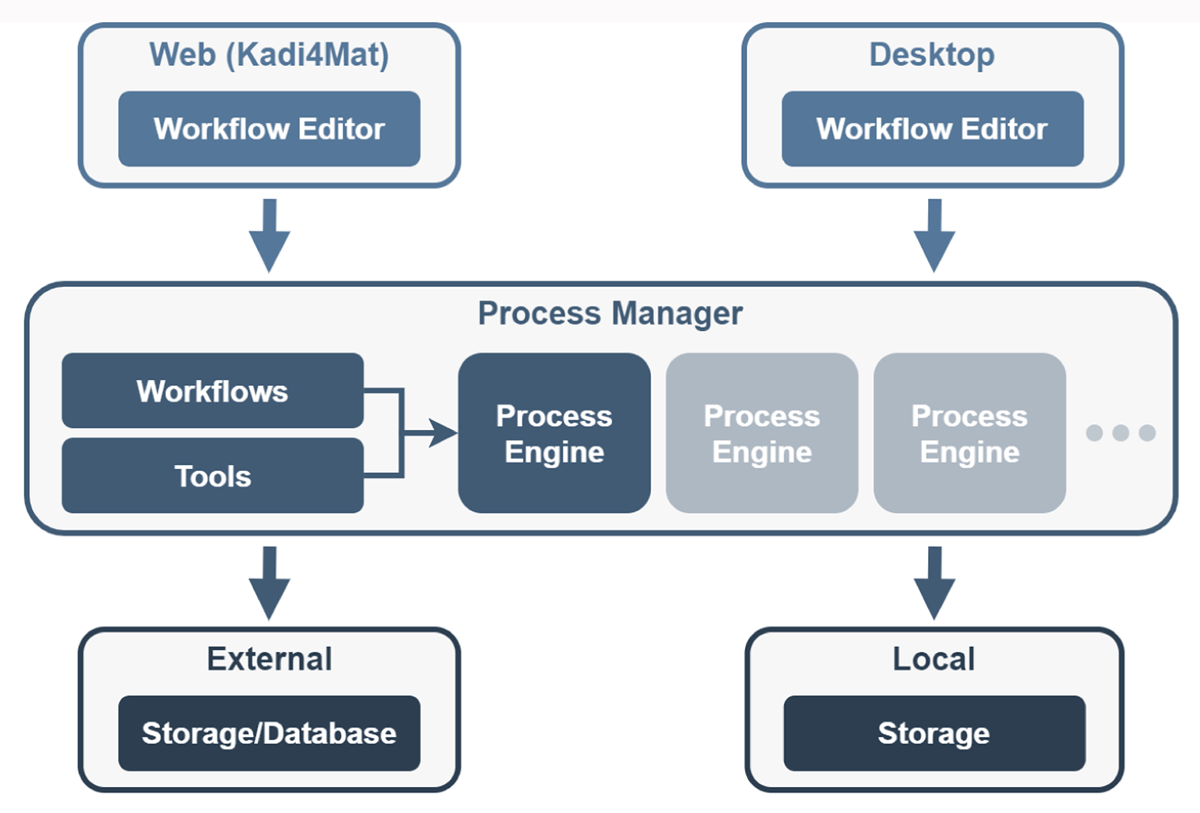
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 2.''' Conceptual overview of the workflow architecture. Each workflow is defined using a graphical editor that is either directly integrated into the web-based interface of Kadi4Mat or locally, with a desktop application. The process manager provides an interface for executing workflows and communicates on behalf of the user with multiple process engines, to which the actual execution of workflows is delegated. The engines are responsible for the actual processing of the different steps, based on the information defined in a workflow. Data and metadata can either be stored externally or locally.</blockquote> | |||
|- | |||
|} | |||
|} | |||
Since the reproducibility of the performed steps is a key objective of the workflows, all meaningful information and metadata can be logged along the way. The logging needs to be flexible in order to accommodate different individual or organizational needs, and as such, it is also part of the workflow itself. Workflows can also be shared with other users, for example, via Kadi4Mat. Manual steps may require interaction during the execution of a workflow, for which the system must prompt the user. In summary, the focus of the ELN component thus points in a different direction than in classic ELNs, with the emphasis on the automation of the steps performed. This aspect in particular is similar to systems such as Galaxy<ref name="AfganTheGal18" />, which focuses on computational biology, or Taverna<ref name="WolstencroftTheTav13">{{cite journal |title=The Taverna workflow suite: designing and executing workflows of Web Services on the desktop, web or in the cloud |journal=Nucleic Acids Research |author=Wolstencroft, K.; Haines, R.; Fellows, D. et al. |volume=41 |issue=W1 |pages=W557–W561 |year=2013 |doi=10.1093/nar/gkt328}}</ref>, a dedicated workflow management system. Nevertheless, some typical features of classic ELNs are also considered in the ELN component, such as the inclusion of handwritten notes. | |||
===Repository=== | |||
In the repository component, data management is regarded as the central element, especially the structured data storage and exchange. An important aspect is the enrichment of data with corresponding descriptive metadata, which is required for its description, analysis, or search. Many repositories, especially those focused on publishing research data, use the metadata schema provided by DataCite<ref name="DataCite43">{{cite web |url=https://schema.datacite.org/meta/kernel-4.3/ |title= DataCite Metadata Schema for the Publication and Citation of Research Data. Version 4.3 |author=DataCite Metadata Working Group |publisher=DataCite e.V |date=16 August 2019}}</ref> and are either directly or heavily based on it. This schema is widely supported and enables the direct publication of data via the corresponding DataCite service. For use cases that go beyond data publications, it is limited in its descriptive power, at the same time. There are comparatively few subject-specific schemas available for engineering and material sciences. Two examples are EngMeta<ref name="SchemberaEngMeta20">{{cite journal |title=EngMeta: Metadata for computational engineering |journal=International Journal of Metadata, Semantics and Ontologies |author=Schembera, B.; Iglezakis, D. |volume=14 |issue=1 |pages=26–38 |year=2020 |doi=10.1504/IJMSO.2020.107792}}</ref> and NOMAD Meta Info.<ref name="GhiringhelliTowards17">{{cite journal |title=Towards efficient data exchange and sharing for big-data driven materials science: metadata and data formats |journal=Computational Materials |author=Ghiringhelli, L.M.; Carbogno, C.; Levchenko, S. et al. |volume=3 |at=46 |year=2017 |doi=10.1038/s41524-017-0048-5}}</ref> The first schema is created ''a priori'' and aims to provide a generic description of computer-aided engineering data, while the second schema is created ''a posteriori'', using existing computing inputs and outputs from the database of the NOMAD repository. | |||
Revision as of 23:43, 22 February 2021
| Full article title | Kadi4Mat: A research data infrastructure for materials science |
|---|---|
| Journal | Data Science Journal |
| Author(s) |
Brandt, Nico; Griem, Lars; Herrmann, Christoph; Schoof, Ephraim; Tosato, Giovanna; Zhao, Yinghan; Zschumme, Philipp; Selzer, Michael |
| Author affiliation(s) | Karlsruhe Institute of Technology, Karlsruhe University of Applied Sciences, Helmholtz Institute Ulm |
| Primary contact | Email: nico dot brandt at kit dot edu |
| Year published | 2021 |
| Volume and issue | 20(1) |
| Article # | 8 |
| DOI | 10.5334/dsj-2021-008 |
| ISSN | 1683-1470 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://datascience.codata.org/articles/10.5334/dsj-2021-008/ |
| Download | https://datascience.codata.org/articles/10.5334/dsj-2021-008/galley/1048/download/ (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
The concepts and current developments of a research data infrastructure for materials science are presented, extending and combining the features of an electronic laboratory notebook (ELN) and a repository. The objective of this infrastructure is to incorporate the possibility of structured data storage and data exchange with documented and reproducible data analysis and visualization, which finally leads to the publication of the data. This way, researchers can be supported throughout the entire research process. The software is being developed as a web-based and desktop-based system, offering both a graphical user interface (GUI) and a programmatic interface. The focus of the development is on the integration of technologies and systems based on both established as well as new concepts. Due to the heterogeneous nature of materials science data, the current features are kept mostly generic, and the structuring of the data is largely left to the users. As a result, an extension of the research data infrastructure to other disciplines is possible in the future. The source code of the project is publicly available under a permissive Apache 2.0 license.
Keywords: research data management, electronic laboratory notebook, repository, open source, materials science
Introduction
In the engineering sciences, the handling of digital research data plays an increasingly important role in all fields of application.[1] This is especially the case, due to the growing amount of data obtained from experiments and simulations.[2] The extraction of knowledge from these data is referred to as a data-driven, fourth paradigm of science, filed under the keyword "data science."[3] This is particularly true in materials science, as the research and understanding of new materials are becoming more and more complex.[4] Without suitable analysis methods, the ever-growing amount of data will no longer be manageable. In order to be able to perform appropriate data analyses smoothly, the structured storage of research data and associated metadata is an important aspect. Specifically, a uniform research data management is needed, which is made possible by appropriate infrastructures such as research data repositories. In addition to uniform data storage, such systems can help to overcome inter-institutional hurdles in data exchange, compare theoretical and experimental data, and provide reproducible workflows for data analysis. Furthermore, linking the data with persistent identifiers enables other researchers to directly reference them in their work.
In particular, repositories for the storage and internal or public exchange of research data are becoming increasingly widespread. In particular, the publication of such data, either on its own or as a supplement to a text publication, is increasingly encouraged or sometimes even required.[5] In order to find a suitable repository, services such as re3data.org[6] or FAIRSharing[7] are available. These services also make it possible to find subject-specific repositories for materials science data. Two well-known examples are the Materials Project[8] and the NOMAD Repository.[9] Indexed repositories are usually hosted centrally or institutionally and are mostly used for the publication of data. However, some of the underlying systems can also be installed by the user, e.g., for internal use within individual research groups. Additionally, this allows full control over stored data as well as internal data exchanges, if this function is not already part of the repository. In this respect, open-source systems are particularly important, as this means independence from vendors and opens up the possibility of modifying the existing functionality or adding additional features, sometimes via built-in plug-in systems. Examples of such systems are CKAN[10], Dataverse[11], DSpace[12], and Invenio[13], where the latter is the basis of Zenodo.[14] The listed repositories are all generic and represent only a selection of the existing open-source systems.[15]
In addition to repositories, a second type of system increasingly being used in experiment-oriented research areas is the electronic laboratory notebook (ELN).[16] Nowadays, the functionality of ELNs goes far beyond the simple replacement of paper-based laboratory notebooks, and can also include aspects such as data analysis, as seen, for example, in Galaxy[17] or Jupyter Notebook.[18] Both systems focus primarily on providing accessible and reproducible computational research data. However, the boundary between unstructured and structured data is increasingly becoming blurred, the latter being traditionally only found in laboratory information management systems (LIMS).[19][20][21] Most existing ELNs are domain-specific and limited to research disciplines such as biology or chemistry.[21] According to current knowledge, a system specifically tailored to materials science does not exist. For ELNs, there are also open-source systems such as eLabFTW[22], sciNote[23], or Chemotion.[24] Compared to the repositories, however, the selection of ELNs is smaller. Furthermore, only the first two mentioned systems are generic.
Thus, generic research data systems and software are available for both ELNs and repositories, which, in principle, could also be used in materials science. The listed open-source solutions are of particular relevance, as they can be adapted to different needs and are generally suitable for use in a custom installation within single research groups. However, both aspects can be a considerable hurdle, especially for smaller groups. Due to a lack of resources, structured research data management and the possibility of making data available for subsequent use is therefore particularly difficult for such groups.[25] What is finally missing is a system that can be deployed and used both centrally and decentrally, as well as internally and publicly, without major obstacles. The system should support researchers throughout the entire research process, starting with the generation and extraction of raw data, up to the structured storage, exchange, and analysis of the data, resulting in the final publication of the corresponding results. In this way, the features of the ELN and the repository are combined, creating a virtual research environment[26] that accelerates the generation of innovations by facilitating collaboration among researchers. In an interdisciplinary field like materials science, there is a special need to model the very heterogeneous workflows of researchers.[4]
For this purpose, the research data infrastructure Kadi4Mat (Karlsruhe Data Infrastructure for Materials Sciences) is being developed at the Institute for Applied Materials (IAM-CMS) of the Karlsruhe Institute of Technology (KIT). The aim of the software is to combine the possibility of structured data storage with documented and reproducible workflows for data analysis and visualization tasks, incorporating new concepts with established technologies and existing solutions. In the development of the software, the FAIR Guiding Principles[27] for scientific data management are taken into account. Instances of the data infrastructure have already been deployed and today show how structured data storage and data exchange are made possible.[28] Furthermore, the source code of the project is publicly available under a permissive Apache 2.0 license.[29]
Concepts
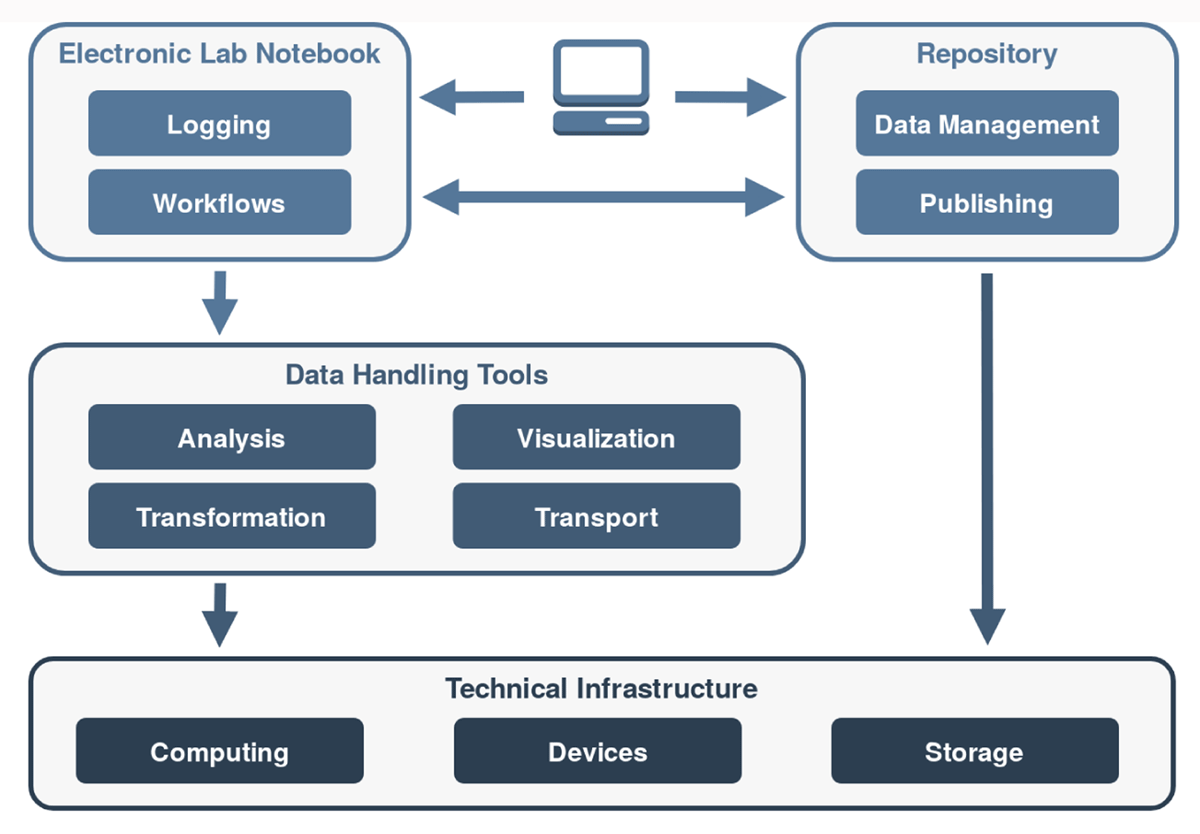
Kadi4Mat is logically divided into the two components—an ELN and a repository—which have access to various tools and technical infrastructures. The components can be used by web- and desktop-based applications, via uniform interfaces. Both a graphical and a programmatic interface are provided, using machine-readable formats and various exchange protocols. In Figure 1, a conceptual overview of the infrastructure of Kadi4Mat is presented.
|
Electronic laboratory notebook
The so-called workflows are of particular importance in the ELN component. A "workflow" is a generic concept that describes a well-defined sequence of sequential or parallel steps, which are processed as automatically as possible. This can include the execution of an analysis tool or the control and data retrieval of an experimental device. To accommodate such heterogeneity, concrete steps must be implemented as flexibly as possible, since they are highly user- and application-specific. In Figure 1, the types of tools shown in the second layer are used as part of the workflows, so as to implement the actual functionality of the various steps. These can be roughly divided into analysis, visualization, transformation, and transportation tasks. In order to keep the application of these tools as generic as possible, a combination of provided and user-defined tools is accessed. From a user’s perspective, it must be possible to provide such tools in an easy manner, while the execution of each tool must take place in a secure and functional environment. This is especially true for existing tools—e.g., a simple MATLAB[30] script—which require certain dependencies to be executed and must be equipped with a suitable interface to be used within a workflow. Depending on their functionality, the tools must in turn access various technical infrastructures. In addition to the use of the repository and computing infrastructure, direct access to devices is also important for more complex data analyses. The automation of a typical workflow of experimenters is only fully possible if data and metadata, created by devices, can be captured. However, such an integration is not trivial due to a heterogeneous device landscape, as well as proprietary data formats and interfaces.[31][32] In Kadi4Mat, it should also be possible to use individual tools separately, where appropriate, i.e., outside a workflow. For example, when using the web-based interface, a visualization tool for a custom data format may be used to generate a preview of a datum that can be directly displayed in a web browser.
In Figure 2, the current concept for the integration of workflows in Kadi4Mat is shown. Different steps of a workflow can be defined with a graphical node editor. Either a web-based or a desktop-based version of such an editor can be used, the latter running as an ordinary application on a local workstation. With the help of such an editor, the different steps or tools to be executed are defined, linked, and, most importantly, parameterized. The execution of a workflow can be started via an external component called "Process Manager." This component in turn manages several process engines, which take care of executing the workflows. The process engines potentially differ in their implementation and functionality. A simple process engine, for example, could be limited to a sequential execution order of the different tasks, while another one could execute independent tasks in parallel. All engines process the required steps based on the information stored in the workflow. With appropriate transport tools, the data and metadata required for each step, as well as the resulting output, can be exported or imported from Kadi4Mat using the existing interfaces of the research data infrastructure. With similar tools, the use of other external data sources becomes possible, and with it the possibility to handle large amounts of data via suitable exchange protocols. The use of locally stored data is also possible when running a workflow on a local workstation.
|
Since the reproducibility of the performed steps is a key objective of the workflows, all meaningful information and metadata can be logged along the way. The logging needs to be flexible in order to accommodate different individual or organizational needs, and as such, it is also part of the workflow itself. Workflows can also be shared with other users, for example, via Kadi4Mat. Manual steps may require interaction during the execution of a workflow, for which the system must prompt the user. In summary, the focus of the ELN component thus points in a different direction than in classic ELNs, with the emphasis on the automation of the steps performed. This aspect in particular is similar to systems such as Galaxy[17], which focuses on computational biology, or Taverna[33], a dedicated workflow management system. Nevertheless, some typical features of classic ELNs are also considered in the ELN component, such as the inclusion of handwritten notes.
Repository
In the repository component, data management is regarded as the central element, especially the structured data storage and exchange. An important aspect is the enrichment of data with corresponding descriptive metadata, which is required for its description, analysis, or search. Many repositories, especially those focused on publishing research data, use the metadata schema provided by DataCite[34] and are either directly or heavily based on it. This schema is widely supported and enables the direct publication of data via the corresponding DataCite service. For use cases that go beyond data publications, it is limited in its descriptive power, at the same time. There are comparatively few subject-specific schemas available for engineering and material sciences. Two examples are EngMeta[35] and NOMAD Meta Info.[36] The first schema is created a priori and aims to provide a generic description of computer-aided engineering data, while the second schema is created a posteriori, using existing computing inputs and outputs from the database of the NOMAD repository.
References
- ↑ Sandfeld, S.; Dahmen, T.; Fischer, F.O.R. et al. (2018). "Strategiepapier - Digitale Transformation in der Materialwissenschaft und Werkstofftechnik". Deutsche Gesellschaft für Materialkunde e.V. https://www.tib.eu/en/search/id/TIBKAT%3A1028913559/.
- ↑ Hey, T.; Trefethen, A. (2003). "Chapter 36: The Data Deluge: An e‐Science Perspective". In Berman, F.; Fox, G.; Hey, T.. Grid Computing: Making the Global Infrastructure a Reality. John Wiley & Sons, Ltd. doi:10.1002/0470867167.ch36. ISBN 9780470867167.
- ↑ Hey, T.; Tansley, S.; Tolle, K. (2009). The Fourth Paradigm: Data-Intensive Scientific Discovery. Microsoft Research. ISBN 9780982544204. https://www.microsoft.com/en-us/research/publication/fourth-paradigm-data-intensive-scientific-discovery/.
- ↑ 4.0 4.1 Hill, J.; Mulholland, G.; Persson, K. et al. (2016). "Materials science with large-scale data and informatics: Unlocking new opportunities". MRS Bulletin 41 (5): 399–409. doi:10.1557/mrs.2016.93.
- ↑ Naughton, L.; Kernohan, D. (2016). "Making sense of journal research data policies". Insights 29 (1): 84–9. doi:10.1629/uksg.284.
- ↑ Pampel, H.; Vierkant, P.; Scholze, F. et al. (2013). "Making Research Data Repositories Visible: The re3data.org Registry". PLoS One 8 (11): e78080. doi:10.1371/journal.pone.0078080.
- ↑ Sansone, S.-A.; McQuilton, P.; Rocca-Serra, P. et al. (2019). "FAIRsharing as a community approach to standards, repositories and policies". Nature Biotechnology 37: 358–67. doi:10.1038/s41587-019-0080-8.
- ↑ Jain, A.; Ong, S.P.; Hautier, G. et al. (2013). "Commentary: The Materials Project: A materials genome approach to accelerating materials innovation". APL Materials 1 (1): 011002. doi:10.1063/1.4812323.
- ↑ Draxl, C.; Scheffler, M. (2018). "NOMAD: The FAIR concept for big data-driven materials science". MRS Bulletin 43 (9): 676–82. doi:10.1557/mrs.2018.208.
- ↑ "CKAN". CKAN Association. https://ckan.org/. Retrieved 19 May 2020.
- ↑ King, G. (2007). "An Introduction to the Dataverse Network as an Infrastructure for Data Sharing". Sociological Methods & Research 36 (2): 173–99. doi:10.1177/0049124107306660.
- ↑ Smith, M.; Barton, M.; Bass, M. et al. (2003). "DSpace: An Open Source Dynamic Digital Repository". D-Lib Magazine 9 (1). doi:10.1045/january2003-smith.
- ↑ "Invenio". CERN. https://invenio-software.org/. Retrieved 19 May 2020.
- ↑ European Organization for Nuclear Research (2013). "Zenodo". CERN. doi:10.25495/7GXK-RD71. https://www.zenodo.org/.
- ↑ Amorim, R.C.; Castro, J.A.; da Silva, J.R. et al. (2017). "A comparison of research data management platforms: Architecture, flexible metadata and interoperability". Universal Access in the Information Society 16: 851–62. doi:10.1007/s10209-016-0475-y.
- ↑ Rubacha, M.; Rattan, A.K.; Hosselet, S.C. (2011). "A Review of Electronic Laboratory Notebooks Available in the Market Today". SLAS Technology 16 (1). doi:10.1016/j.jala.2009.01.002.
- ↑ 17.0 17.1 Afgan, E.; Baker, D.; Batut, B. et al. (2018). "The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update". Nucleic Acids Research 46 (W1). doi:10.1093/nar/gky379.
- ↑ Kluyver, T.; Ragan-Kelley, B.; Pérez, F. et al. (2016). "Jupyter Notebooks—A publishing format for reproducible computational workflows". In Loizides, F.; Schmidt, B.. Positioning and Power in Academic Publishing: Players, Agents and Agendas. IOS Press. pp. 87–90. doi:10.3233/978-1-61499-649-1-87.
- ↑ Bird, C.L.; Willoughby, C.; Frey, J.G. (2013). "Laboratory notebooks in the digital era: The role of ELNs in record keeping for chemistry and other sciences". Chemical Society Reviews 42 (20): 8157–8175. doi:10.1039/C3CS60122F.
- ↑ Elliott, M.H. (2009). "Thinking Beyond ELN". Scientific Computing 26 (6): 6–10. Archived from the original on 20 May 2011. https://web.archive.org/web/20110520065023/http://www.scientificcomputing.com/articles-IN-Thinking-Beyond-ELN-120809.aspx.
- ↑ 21.0 21.1 Taylor, K.T. (2006). "The status of electronic laboratory notebooks for chemistry and biology". Current Opinion in Drug Discovery and Development 9 (3): 348–53. PMID 16729731.
- ↑ Carpi, N.; Minges, A.; Piel, M. (2017). "eLabFTW: An open source laboratory notebook for research labs". Journal of Open Source Software 2 (12): 146. doi:10.21105/joss.00146.
- ↑ "SciNote". SciNote LLC. https://www.scinote.net/. Retrieved 21 May 2020.
- ↑ Tremouilhac, P.; Nguyen, A.; Huang, Y.-C. et al. (2017). "Chemotion ELN: An open source electronic lab notebook for chemists in academia". Journal of Cheminformatics 9: 54. doi:10.1186/s13321-017-0240-0.
- ↑ Heidorn, P.B. (2008). "Shedding Light on the Dark Data in the Long Tail of Science". Library Trends 57 (2): 280–99. doi:10.1353/lib.0.0036.
- ↑ Carusi, A.; Reimer, T.F. (17 January 2010). "Virtual Research Environment Collaborative Landscape Study". JISC. doi:10.25561/18568. http://hdl.handle.net/10044/1/18568.
- ↑ Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J. et al. (2016). "The FAIR Guiding Principles for scientific data management and stewardship". Scientific Data 3: 160018. doi:10.1038/sdata.2016.18. PMC PMC4792175. PMID 26978244. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4792175.
- ↑ "Kadi4Mat". Karlsruhe Institute of Technology. https://kadi.iam-cms.kit.edu/. Retrieved 30 September 2020.
- ↑ Brandt, N.; Griem, L.; Hermann, C. et al. (2020). "IAM-CMS/kadi: Kadi4Mat (Version 0.6.0)". Zenodo. doi:10.5281/zenodo.4507826.
- ↑ "MATLAB". MathWorks. https://www.mathworks.com/products/matlab.html. Retrieved 19 January 2021.
- ↑ Hawker, C.D. (2007). "Laboratory Automation: Total and Subtotal". Clinics in Laboratory Medicine 27 (4): 749–70. doi:10.1016/j.cll.2007.07.010.
- ↑ Potthoff, J.; Tremouilhac, P.; Hodapp, P. et al. (2019). "Procedures for systematic capture and management of analytical data in academia". Analytica Chimica Acta: X 1: 100007. doi:10.1016/j.acax.2019.100007.
- ↑ Wolstencroft, K.; Haines, R.; Fellows, D. et al. (2013). "The Taverna workflow suite: designing and executing workflows of Web Services on the desktop, web or in the cloud". Nucleic Acids Research 41 (W1): W557–W561. doi:10.1093/nar/gkt328.
- ↑ DataCite Metadata Working Group (16 August 2019). "DataCite Metadata Schema for the Publication and Citation of Research Data. Version 4.3". DataCite e.V. https://schema.datacite.org/meta/kernel-4.3/.
- ↑ Schembera, B.; Iglezakis, D. (2020). "EngMeta: Metadata for computational engineering". International Journal of Metadata, Semantics and Ontologies 14 (1): 26–38. doi:10.1504/IJMSO.2020.107792.
- ↑ Ghiringhelli, L.M.; Carbogno, C.; Levchenko, S. et al. (2017). "Towards efficient data exchange and sharing for big-data driven materials science: metadata and data formats". Computational Materials 3: 46. doi:10.1038/s41524-017-0048-5.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original article lists references in alphabetical order; however, this version lists them in order of appearance, by design.