Journal:Laboratory automation, informatics, and artificial intelligence: Current and future perspectives in clinical microbiology
| Full article title | Laboratory automation, informatics, and artificial intelligence: Current and future perspectives in clinical microbiology |
|---|---|
| Journal | Frontiers in Cellular and Infection Microbiology |
| Author(s) | Mencacci, Antonella; De Socio, Guiseppe V.; Pirelli, Eleonora; Bondi, Paola; Cenci, Elio |
| Author affiliation(s) | University of Perugia, Perugia General Hospital |
| Primary contact | Email: antonella at mencacci at unipg dot it |
| Year published | 2023 |
| Volume and issue | 13 |
| Article # | 1188684 |
| DOI | 10.3389/fcimb.2023.1188684 |
| ISSN | 2235-2988 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fcimb.2023.1188684/full |
| Download | https://www.frontiersin.org/articles/10.3389/fcimb.2023.1188684/pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Clinical diagnostic laboratories produce one product—information—and for this to be valuable, the information must be clinically relevant, accurate, and timely. Although diagnostic information can clearly improve patient outcomes and decrease healthcare costs, technological challenges and laboratory workflow practices affect the timeliness and clinical value of diagnostics. This article will examine how prioritizing laboratory practices in a patient-oriented approach can be used to optimize technology advances for improved patient care.
Keywords: laboratory automation, artificial intelligence, informatics, laboratory workflow, Kiestra, WASPLab
Introduction
Patterns of infectious diseases have changed dramatically: patients are frequently immunocompromised and often have complicating comorbidities; infections with multi-drug-resistant organisms (MDRO) are a global problem; and new antibiotics are available, but it is mandatory to preserve their efficacy. It is estimated that at least 700,000 people die worldwide every year with infections caused by MDRO, and it is predicted that by 2050, 10 million deaths might occur due to these organisms. [O’Neill, 2016] Administration of rapid, broad-spectrum empiric therapy is essential to improve patient outcome [Levy et al., 2018], but this is often inappropriate. [Kumar et al., 2009; Zilberberg et al., 2017] For example, meta-analysis assessing the impact of antibiotic therapy on Gram-negative sepsis showed that inappropriate therapy was associated with 3.3-fold increased risk of mortality, longer hospitalization, and higher costs. [Raman et al., 2015] Thus, rapid, accurate diagnostics are critical for the selection of the most appropriate therapy.
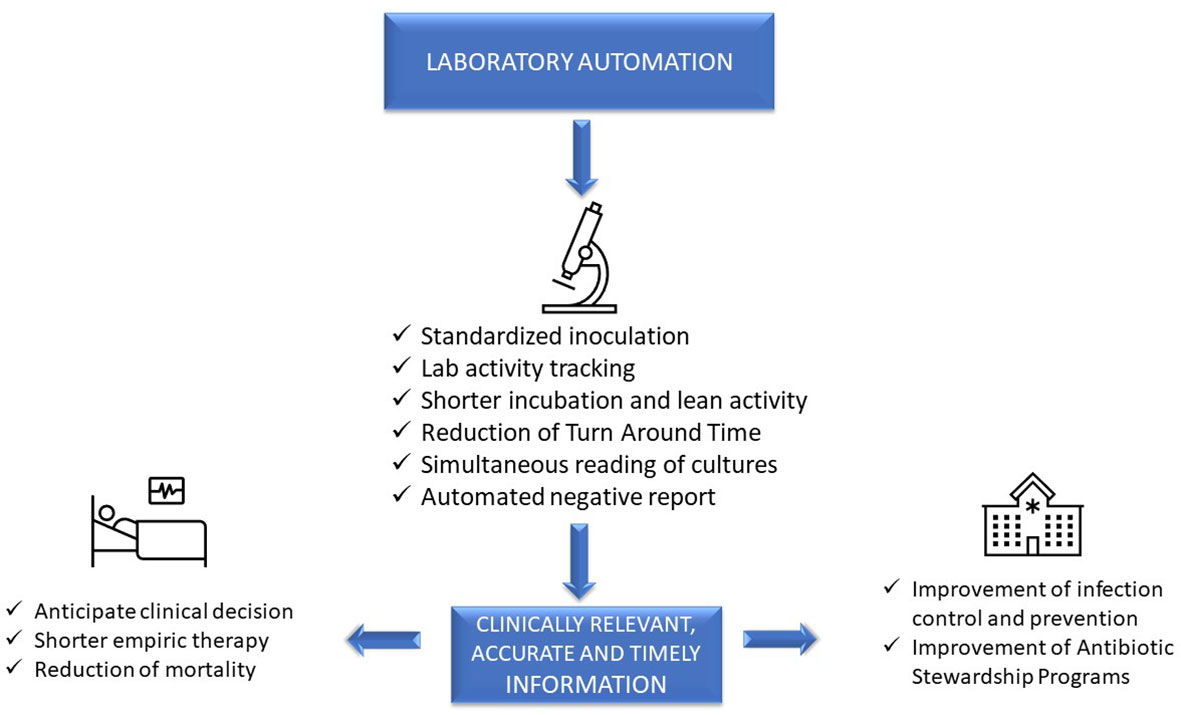
Advanced, sophisticated technologies such as mass spectrometry and molecular diagnostics are rapidly changing our ability to diagnose infections [Trotter et al., 2019], although they should be viewed as complementary to traditional growth-based diagnostics. Laboratory automation and intelligent applications of of informatics also have a transformative impact of microbiology diagnostics. These tools have the potential to accelerate clinical decision-making and positively impact the management of infections, improve patient outcome, and facilitate diagnostic and antimicrobial stewardship (AS) programs. [Messacar et al., 2017] However, it is a challenge for clinical microbiologists to implement these technologies because it requires changing well-established workflow practices. This paper will focus on the impact of automation and informatics combined with workflow changes on laboratory, patient, and hospital management (Figure 1).
|
Impact on laboratory management
In clinical microbiology, the term “total laboratory automation” (TLA) is used to describe the automation of the entire diagnostic workflow, from inoculation of the agar plates to incubation, reading of culture results, identification (ID), and antimicrobial susceptibility testing (AST). All these steps in a conventional laboratory are performed manually, usually according to a sample-centered approach. At present, two laboratory automation systems are available: the BD Kiestra system (Becton Dickinson, Sparks, MD) and the WASPLab system (Copan Diagnostics, Murrieta, CA). [Croxatto et al., 2016] We discuss these systems within the context of this workflow and then address other laboratory automation tools, as well artificial intelligence (AI).
Inoculation
Quality and precision of inoculation are improved by automation. Instruments work in a standardized and consistent mode, not achievable with a manual procedure, and independent of operator variability. Indeed, laboratory automation allows better isolation of colonies compared to manual inoculation, with decreased need of subcultures for follow-up work, mainly AST, resulting in a more rapid report. [Croxatto et al., 2015; Burckhardt, 2018] It was found that WASP automated streaking of urines using a sterile loop was superior to manual streaking, yielding a higher number of single colonies and of detected morphologies, species, and pathogens. [Quiblier et al., 2016] The BD Kiestra system, based on a rolling magnetic bead streaking technology, has been shown to improve the accuracy of quantitative culture results and the recovery of discrete colonies from polymicrobial samples, compared to manual and automated WASP streaking. [Croxatto et al., 2015; Iversen et al., 2016] This implies a reduction of bacterial subcultures to perform ID and AST, thus shortening time to results, as evidenced for urines [Croxatto et al., 2017] and both methicillin-resistant Staphylococcus aureus (MRSA) and carbapenem-resistant Enterobacterales screening samples. [Cheng et al., 2020]
Incubation
Closed incubators with digital imaging of cultures allow more rapid growth than conventional incubators that are opened frequently throughout the day. Moreover, in TLA, plates are fully tracked as long as they stay within the system, so that it is possible to define by hours and minutes incubation times and plate examination, in contrast with the traditional system in which incubation times are defined in days. Burckhardt et al. showed that first growth of MRSA, multi-drug-resistant (MDR) Gram-negative bacteria, and vancomycin-resistant enterococci (VRE) on selective chromogenic plates was visible as early as after four hours of inoculation, although the bacterial mass was not sufficient for follow-up work. [Burckhardt et al., 2019] Also, growth of Escherichia coli, Pseudomonas aeruginosa, Enterococcus faecalis, and S. aureus on chromogenic plates was three to four hours faster in the automated system than in the classic system. [Moreno-Camacho et al., 2017] Implementation of BD Kiestra TLA significantly improved turnaround times (TAT) for positive and negative urine cultures. [Theparee et al., 2017] Similarly, WASPLab automation enabled a reduction of the culture reading time for different specimens without affecting performances. [Cherkaoui et al., 2019] However, minimum incubation times for each type of specimen, for a timely and accurate positive or negative report, are not yet defined, and additional studies are needed.
Reading
The Kiestra laboratory automation system, through a real-time dashboard, times tasks as they are scheduled. Thus, each technician perfectly knows when the culture plates will be ready for reading and when follow-up work can be performed. This strongly facilitates laboratory workflow management, avoiding wasted time and allowing results to be delivered to the clinician as soon as possible. In addition, while in the classical system plates are read one by one, digital reading allows simultaneous viewing of all the plate images from the same sample, and even of different samples from the same patient. This greatly facilitates and speeds up the interpretation of culture results, either for monomicrobial or polymicrobial infections.
ID and AST
The implementation of TLA in clinical microbiology has leveraged the advancement brought by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS). [Thomson and McElvania, 2019; Cherkaoui and Schrenzel, 2022] Furthermore, Copan’s TLA has recently integrated an automated device (Colibri) that can reproducibly prepare the MALDI target for microbial identification. A recent study conducted by Cherkaoui et al. established that the WASPLab coupled to MALDI-TOF/MS significantly reduces the TAT for positive blood cultures. [Cherkaoui et al., 2023] Similarly, the BD Kiestra IdentifA/SusceptA, a prototype for automatic colony picking, bacterial suspension preparation, MALDI-TOF target plates spotting, and Phoenix M50 AST panel preparation, exhibited high ID and AST performances. [Jacot et al., 2021] In particular, the IdentifA showed excellent identification rates for Gram-negative bacteria, outperforming manual processing for Enterobacterales identification [Jacot et al., 2021], but not for streptococci, coagulase-negative staphylococci (CoNS), and yeasts. [Jacot et al., 2021]
Finally, an automated solution for disk diffusion AST was developed and integrated with the Copan WASPLab system. It prepares inoculum suspensions, inoculates culture media plates, dispenses appropriate antibiotic disks according to predefined panels, transports the plates to the incubators, takes digitalized images of the media plates, and measures and interprets the inhibition zones’ diameters. Cherkaoui et al.—evaluating 718 bacterial strains, including S. aureus, CoNS, E. faecalis, Enterococcus faecium, P. aeruginosa, and different species of Enterobacterales—found 99.1% overall categorical agreement between this automated AST and Vitek2. [Cherkaoui et al., 2021]
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original article lists references alphabetically; they are listed by order of appearance for this version, by design.