Journal:Digital pathology and anatomic pathology laboratory information system integration to support digital pathology sign-out
| Full article title | Digital pathology and anatomic pathology laboratory information system integration to support digital pathology sign-out |
|---|---|
| Journal | Journal of Pathology Informatics |
| Author(s) |
Guo, Huazhang; Birsa, Joe; Farahani, Navid; Hartman, Douglas J.; Piccoli, Anthony; O'Leary, Matthew; McHugh, Jeffrey; Nyman, Mark; Stratman, Curtis; Kvarnstrom, Vanja; Yousem, Samuel; Pantanowitz, Liron |
| Author affiliation(s) | University of Pittsburgh Medical Center and Omnyx, LLC |
| Primary contact | Email: Available w/ login |
| Year published | 2016 |
| Volume and issue | 7 |
| Page(s) | 23 |
| DOI | 10.4103/2153-3539.181767 |
| ISSN | 2153-3539 |
| Distribution license | Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported |
| Website | http://www.jpathinformatics.org |
| Download | http://www.jpathinformatics.org/temp/JPatholInform7123-3954163_105901.pdf (PDF) |
Abstract
Background: The adoption of digital pathology offers benefits over labor-intensive, time-consuming, and error-prone manual processes. However, because most workflow and laboratory transactions are centered around the anatomical pathology laboratory information system (APLIS), adoption of digital pathology ideally requires integration with the APLIS. A digital pathology system (DPS) integrated with the APLIS was recently implemented at our institution for diagnostic use. We demonstrate how such integration supports digital workflow to sign-out anatomical pathology cases.
Methods: Workflow begins when pathology cases get accessioned into the APLIS (CoPathPlus). Glass slides from these cases are then digitized (Omnyx VL120 scanner) and automatically uploaded into the DPS (Omnyx; Integrated Digital Pathology (IDP) software v.1.3). The APLIS transmits case data to the DPS via a publishing web service. The DPS associates scanned images with the correct case using barcode labels on slides and information received from the APLIS. When pathologists remotely open a case in the DPS, additional information (e.g. gross pathology details, prior cases) gets retrieved from the APLIS through a query web service.
Results: Following validation of this integration, pathologists at our institution have signed out more than 1000 surgical pathology cases in a production environment. Integration between the APLIS and DPS enabled pathologists to review digital slides while simultaneously having access to pertinent case metadata. The introduction of a digital workflow eliminated costly manual tasks involving matching of glass slides and avoided delays waiting for glass slides to be delivered.
Conclusion: Integrating the DPS and APLIS were instrumental for successfully implementing a digital solution at our institution for pathology sign-out. The integration streamlined our digital sign-out workflow, diminished the potential for human error related to matching slides, and improved the sign-out experience for pathologists.
Keywords: Digital pathology, integration, interface, laboratory information system, whole slide imaging
Introduction
Currently, there are two key information technology (IT) building blocks available to anatomic pathology laboratories that intend to implement a digital surgical pathology solution. They are the anatomic pathology laboratory information system (APLIS) and digital pathology system (DPS) that uses whole slide images (WSIs). The APLIS manages most routine surgical pathology workflow in practice and has been universally adopted in anatomic pathology laboratories.[1] The APLIS successfully handles many functions such as managing worklists, tracking assets, generating pathology reports and billing. Within the APLIS, surgical pathology sign-out is largely computerized (e.g., electronic sign-out) but remains semi-digital when using glass slides. Several of the steps in current iterations of anatomical pathology sign-out workflow that remain nondigital include histotechnologists manually sorting and matching glass slides to corresponding accessioned cases in the APLIS, and then manually delivering them to pathologists to review using a traditional light microscope. These manual tasks are not only time- and labor-intensive but also error-prone and costly (e.g., expense of using slide trays and courier services). If the histology laboratory and pathologist's office are in different locations, physically delivering glass slides may further increase turnaround time. Fortunately, many shortcomings of these manual processes can be addressed by a DPS.[2][3][4]
The DPS is a relatively new information system developed primarily to manage WSI. The DPS is somewhat analogous to a radiology picture archiving and communication system (PACS).[5][6] Workflow in a DPS typically starts with the creation of a WSI and subsequent storage of these images in a digital image repository (e.g., server). End-users such as pathologists need to access these archived digital slides for viewing and if necessary, share them with others, perform annotation and/or image analysis. In order for clinical work such as digital sign-out for primary diagnosis or teleconsultation to be practical and efficient, WSI needs to be associated with relevant case metadata (e.g., patient demographics, gross pathology findings, prior case diagnoses). This can be accomplished by integrating the APLIS and DPS, which in turn should help streamline digital sign-out workflow, minimize redundant and error-prone manual work, reduce delivery costs of glass slides, and take advantage of computer-assisted quantification and diagnostic algorithms.
At our institution, we implemented a DPS and initiated a multi-year plan to transition our anatomical pathology practice to a digital sign-out workflow. The key to our implementation plan was to integrate the DPS with our legacy APLIS. This article describes our technical approach to APLIS-DPS integration and highlights the benefits of this integration.
Methods
Information systems
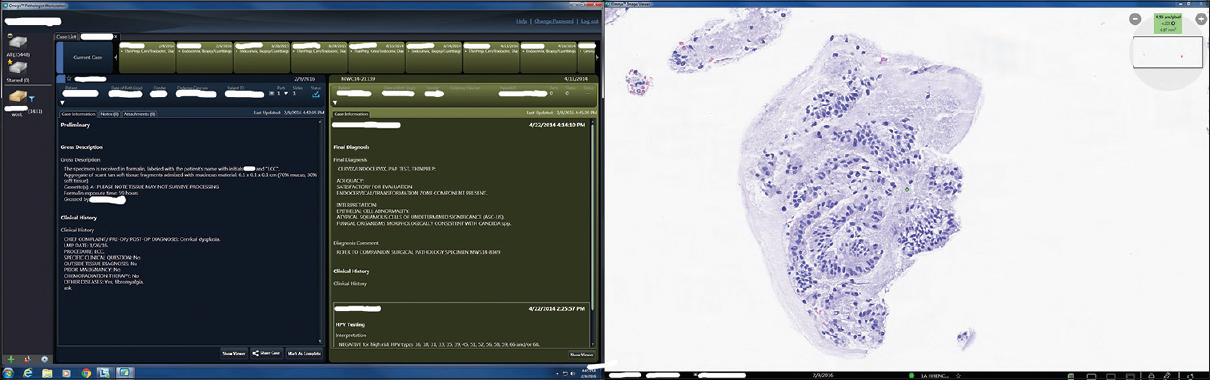
Our APLIS (CoPathPlus, Cerner Corp., Kansas City, MO, USA) has been used for many years to manage accessioning of anatomical pathology cases, handle specimens (e.g., histology, stains, barcode tracking), reporting, and billing. The CoPathPlus database (Sybase V15) is run on a Windows Server (2012). Omnyx IDP (Omnyx Inc., Pittsburgh, PA, USA) was the DPS selected to manage WSI digitization, digital case workflow, and viewing of images. The DPS hardware was comprised of Omnyx VL120 scanners for image acquisition, histology workstations for image quality review and digital case assembly, and pathologist workstations for end-user case management and digital image review. The Omnyx IDP database (Microsoft SQL Server 2012), workflow and diagnostic archive servers were run on a Windows Server (2012) virtual machine environment. The Omnyx pathologist workstation includes two side-by-side displays (HP Z24s Generic PnP monitors with 3840 × 2160 resolution), where the left display showed text-based case-level information and the right screen was dedicated largely to displaying the WSI.
Integration
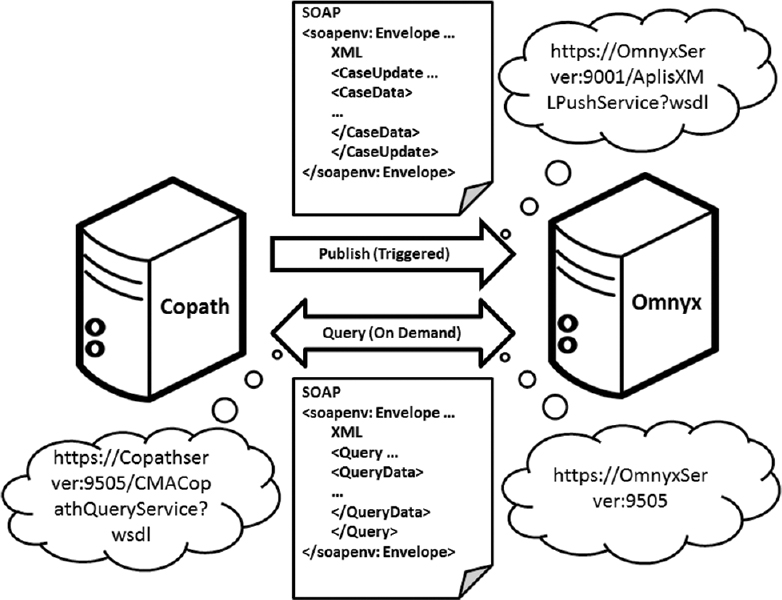
The help of both vendors was solicited to participate in establishing an interface between the APLIS and DPS for our institution. A back-end, two-stage information transfer mechanism was employed using Windows Communication Foundation web services to integrate the DPS with our APLIS [Figure 1]. The first interactive stage occurs within the APLIS, triggered by certain events such as when a case gets accessioned, a case is updated, case histology gets added/changed/deleted, and when a case gets signed out. At this time, when a case is accessioned into the APLIS, the APLIS acting as a web client sends notifications containing case and histology data (e.g., patient name/identifiers/date of birth/gender, case accession number/date/priority/ordering provider identifier/primary pathologist identifier, part/block/slide data, slide status, etc.) to the DPS, which hosts the receiving web service. After receiving case data (e.g., accession number, ordering physician, etc.), patient data (e.g., name, date of birth), and histology data (e.g., parts/blocks/slides), the DPS stores this metadata within its database. When the WSIs are ingested, they can be associated with all of these case-related data based on the unique slide identification (i.e., barcode assigned by the APLIS and read by the DPS on the slide label). The second interactive stage involves a query service request initiated by the DPS, triggered by the end-user opening a case in the DPS. Hence, when a pathologist opens a case in the DPS this action acts as a web client that sends a query to the APLIS, which hosts the query web service. After receiving the demanding query from the DPS, the APLIS responds with additional case-related information (e.g., updated case data, clinical data, gross description, and current and prior case detail data) back to the DPS.
|
To ensure reliable communication between the DPS and APLIS, we implemented an alarm into each system. In the DPS, after sending the APLIS a query message the alarm system gets triggered by failing to receive a response message from the APLIS. Similarly, in the APLIS, after sending the DPS a publish message the alarm system was designed to be triggered by failing to receive an acknowledgment message from the DPS. If the alarms were triggered, our IT team would engage and start troubleshooting. To ensure point-to-point security, hypertext transfer protocol secure (HTTPS) was used to make connections between the DPS and APLIS.
Validation
After the aforementioned interface was built and verified in the "test system," we went live with our DPS "production system" for clinical sign-out on August 2015. Clinical validation involved training technologists to scan glass slides and manage digital slides (e.g. perform quality checks, assign cases), as well as one-on-one training of pathologists to view, interpret, and sign-out digital cases using the Omnyx workstation. Surgical pathology cases selected for our prospective validation study included only small biopsies of gastrointestinal, gynecological, genitourinary, dermatologic, and pediatric pathology at our academic hospitals. Autopsy slides were also included. These were routine cases (not consults cases). Feedback was solicited from all end-users about technical, workflow and/or image quality issues.
Results
The web service middleware used to interface our APLIS with the DPS permitted all case data from the APLIS, that was relevant for a pathologist to sign-out a case, to be simultaneously presented alongside WSI images in the DPS. A screenshot from a pathologist's workstation shown in [Figure 2] illustrates user configurable case information on the left screen and the right screen showing the matched WSI image for that case. There were 24 pathologists trained to use the Omnyx system and 1336 validation cases successfully signed-out in a 34 week period. Diagnostic concordance findings with comparative glass slide review will be communicated in a subsequent publication, as this is out of scope for this paper.
|
The validation study, performed at two of our pilot hospitals, showed that average slide turnaround time (see definition and explanation below) was reduced by at least 1 h per case, with a potential cost saving of around $60,000 per year specifically for avoiding manual slide delivery and buying slide trays. For many years, our institution has relied on using a courier service to deliver newly prepared glass slides from our central histology laboratory to distributed hospitals. For the two hospitals where we initially did the validation study, the courier service picks up and delivers slides 5-6 times (approximately every 2 h) each work day. Slide turnaround time is defined as the time elapsed between prepared glass slides being ready in the histology laboratory for distribution until they are available for pathologists to be viewed. For the digital workflow, the equivalent turnaround time is from the time slides are scanned until they are electronically transmitted to pathologists. Because our courier performs glass slide deliveries every 2 h, on average, there is a 1 h delay for glass slides to be reviewed by a pathologist compared to the digital workflow. The cost of delivering slides to each hospital is approximately $18 per courier trip, which could be eliminated with a digital workflow. In addition, our central histology laboratory would no longer need to purchase 600 slide trays per year (with 1200-1500 in circulation), where the cost of each tray is $10. The financial return of other direct and indirect benefits (e.g., core laboratory facility with centralization of services, going paperless, etc.), was not included in this article, as they have been previously published by our institution.[7]
Two issues surfaced after the web services-based integration went live. The first issue was related to delayed message delivery. While the Publish services proved to be robust, the Omnyx query services were inconsistent resulting in occasional delay. The reason was that all Omnyx publish and query messages were lined up in a single queue and the processing sequence for the queue was first-in-first-out. The time sensitive query messages were lined behind many background publish messages. Over time, the accumulation of background publish messages resulted in a performance degradation of the Microsoft Queue process module and caused significant delay for time sensitive query messages to be processed. A temporary solution was implemented involving rebooting the system at midnight to restart the Microsoft queue process module. As a result, redesign of the queue messaging system is planned for the next version of the system. The second issue concerned the status of completed cases. The initial design was to make a case status change from incomplete to complete if all of the slides were scanned into the DPS. However, in certain situations pathologists preemptively ordered blank slides that do not get digitized, but may eventually only use some of them later at which time they do need to be scanned. In cases where unused blank slides are not stained and scanned, the case status will never change from incomplete to complete. One strategy in which this was addressed was to classify slides into viewable and nonviewable slides in the APLIS, and have the case status not depend on nonviewable slides.
The pathologists involved in the validation study were asked for feedback about the integrated digital workflow. Based on their responses, they were happy with (1) the fact that they did not need to wait for the courier to deliver their glass slides, and that digital slides were available for review immediately after scanning; (2) pertinent case information (e.g., clinical history, gross description) was made available to them while reviewing digital slides, avoiding having them log into to the separate LIS to obtain this information; and (3) the response of the image server was prompt while navigating the images. One desired feature, not yet available, was to be able to view gross pictures in the digital pathology workstation.
Discussion
Integrating heterogeneous computer information systems is a complex task involving many components and established standards. To promote and facilitate integration, Integrating the Healthcare Enterprise (IHE) published a technical framework for the implementation of integration in pathology laboratories.[8] Although our integration project was geminated and designed before the IHE publication, IHE terminology can nonetheless be used to describe our integration implementation. According to IHE, in our institution, the APLIS functions as an order. The DPS functions as an acquisition modality, image manager, image archive, image display, and evidence creator. Web service message exchange is accordingly handling transactions between the order filler and image manager.
Only a few pathology laboratories have reported going fully digital.[9][10] These authors stress that optimization of digital pathology workflow requires communication between disparate systems. For example, the pathology department at University Medical Center in Utrecht, The Netherlands implemented a connection between their internal reporting system and image management system to display digital slides linked to every case number.[9] At Linköping University Hospital in Sweden the pathology department used middleware (Picsara software) to connect their scanned images with cases in the laboratory information system.[10] At Washington University School of Medicine in St. Louis, USA the Pathology Department pursued a model of "one-stop-shopping" by developing an interface between their laboratory information system (Cerner CoPathPlus) and imaging software (Aperio Spectrum). Their interface took approximately 11 months to develop, including 2 months to implement within their existing workflow.[11]
Ideally, there would be no need for integration if one vendor offered a single information system with all the functionality currently available in the APLIS and DPS. However, the common scenario as illustrated by our shared experience is where pathology laboratories have an existing legacy APLIS and need to implement a new DPS supported by a different vendor. Users, therefore, will most likely need to integrate two independent information systems if they want to take advantage of both an APLIS and DPS. Working with Omnyx and Cerner vendors, we implemented a DPS-driven interface between our APLIS and the DPS. Although the DPS and APLIS were initially agnostic to each other's internal database structure, we were able to successfully integrate these two systems using web services. Having pertinent metadata such as gross descriptions and prior cases accessible at the time of viewing a WSI facilitated workflow and enabled pathologists to sign-out cases without glass slides and paperwork.
Integration of healthcare information systems can be undertaken on the back- and/or front-end.[12] Back-end refers to the database or server-side, and front-end to the user or client-side. We opted for back-end integration of the APLIS and DPS. Both systems communicate with each other on the server-side through publish and query web services. Albeit that a back-end solution can integrate all of the data in the databases of heterogeneous applications, it is expensive and lengthy to develop. For front-end integration, the solution needs to intercept interactions between different information systems and act as a middle-tier application server, transparently marshaling, managing, and directing inter-application requests and responses.[12] However, the functionality of front-end integration relies on the application programing interface (API) provided by individual information systems. Few legacy healthcare information systems actually provide an appropriate API for front-end integration. Nevertheless, we are in the process of also developing front-end integration using vergence (Caradigm, LLC) software to facilitate single sign-on and case context sharing between the APLIS and DPS.
Web services is one of several standardized ways of integrating disparate systems.[13] Web services use Extensible Markup Language (XML), simple object access protocol (SOAP), and web service definition language (WSDL) open standards over HTTP/HTTPS. XML is used to tag data, SOAP is used to transfer data, and WSDL is used for describing services. Web services allow disparate systems to communicate data without intimate knowledge of each other's internal data structure. It employs client/server network architecture for communication between disparate information systems. However, unlike traditional client/server architectures, such as internet browser/internet web server architecture, web services do not provide the user with a graphic user interface. Instead, a programmatic interface across a network is used to share data between disparate information systems.
Although we successfully integrated the APLIS and DPS using web service technology, web services are far from a perfect solution in healthcare integration. It has its disadvantages, such as reusability and the cost associated with development and maintenance. Currently, HL7 v2 is the most widely used standard to integrate heterogeneous healthcare information systems.[14] The popularity of HL7 v2 implies that integration interface middleware built using HL7 v2 will have a better chance of being reused by other customers with a greater likelihood of cost sharing by all customers. Our group is investigating other integration strategies such as adding a mirth based interface engine. A mirth interface engine will decouple the interface implementation on both the APLIS and DPS sides, providing flexibility to choose any integration standard including HL7 v2, any upcoming new HL7 standard or fast healthcare interoperability resources.
There were two reasons why information transfer was divided into two stages in the interface we developed. The first was to reduce the unnecessary transfer of information from the APLIS to the DPS. For example, there was no need to transfer over "gross only" cases in the APLIS when no slides were generated. The second reason was to ensure that case-related information reflected the most up-to-date details from the APLIS (e.g., re-assigning of a case to another pathologist). The HTTPS protocol was selected for secure communication of patient healthcare information over our network. HTTPS not only provided authentication of the APLIS and DPS web servers but also provided bidirectional encryption of communications between them. Therefore, communications between the APLIS and DPS cannot be read or forged by a third party.
The APLIS-DPS integration enabled subspecialty pathologists at our institution to review digital slides on one monitor while simultaneously viewing pertinent case metadata such as clinical information and gross pathology findings. In addition, data including WSI from prior cases could be easily reviewed when needed. The benefits of transitioning to a digital workflow include: (1) More efficient and streamlined workflow (e.g., searching for relevant case data in the APLIS), (2) minimizing errors by eliminating manual processes (e.g., matching slides to cases), and (3) reducing turnaround time (e.g., electronically transferring WSI images rather than physically delivering glass slides by courier). Fitting into the radiology PACS maturity model[15], our current integration is at maturity level 2-3 (out of 5 levels). Integrating the APLIS and DPS is a significant step forward toward achieving a comprehensive digital sign-out workflow. However, there are several aspects we intend to address to improve upon integration. This includes establishing bidirectional data flow between the APLIS and DPS so that pathologists can easily perform tasks (e.g., ordering immunohistochemistry stains) and directly provide diagnostic pathology reports from the DPS. Furthermore, we are aware that to optimize the pathology cockpit (or digital dashboard) it is important to also integrate clinical information and radiology images from other electronic medical records.[16][17] In the near future, perhaps interoperability may also be facilitated by widespread adoption of standards such as DICOM.[8]
Acknowledgements
This study was presented in part at the USCAP annual meeting in Seattle, March 12-18, 2016.
Financial support and sponsorship
Nil
Conflicts of interest
UPMC is a part owner of Omnyx. Dr. Pantanowitz and Dr. Guo are consultants for Omnyx.
References
- ↑ Park, S.L.; Pantanowitz, L.; Sharma, G.; Parwani, A.V. (2012). "Anatomic pathology laboratory information systems: A review". Advances in Anatomic Pathology 19 (2): 81–96. doi:10.1097/PAP.0b013e318248b787. PMID 22313836.
- ↑ Park, S.L.; Pantanowitz, L.; Parwani, A.V. (2012). "Digital imaging in pathology". Clinics in Laboratory Medicine 32 (4): 557–84. doi:10.1016/j.cll.2012.07.006. PMID 23078660.
- ↑ Cornish, T.C.; Swapp, R.E.; Kaplan, K.J. (2012). "Whole-slide imaging: Routine pathologic diagnosis". Advances in Anatomic Pathology 19 (3): 152–9. doi:10.1097/PAP.0b013e318253459e. PMID 22498580.
- ↑ Fine, J.L. (2014). "21st century workflow: A proposal". Journal of Pathology Informatics 5 (1): 44. doi:10.4103/2153-3539.145733. PMC PMC4260324. PMID 25535592. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4260324.
- ↑ Morelli, S.; Grigioni, M.; Giovagnoli, M.R.; Balzano, S.; Giansanti, D. (2010). "Picture archiving and communication systems in digital cytology". Annali dell'Istituto Superiore di Sanità 46 (2): 130–7. doi:10.4415/ANN_10_02_05. PMID 20567063.
- ↑ Tuominen, V.J.; Isola, J. (2010). "Linking whole-slide microscope images with DICOM by using JPEG2000 interactive protocol". Journal of Digital Imaging 23 (4): 454–62. doi:10.1007/s10278-009-9200-1. PMC PMC2896636. PMID 19415383. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2896636.
- ↑ Ho, J.; Ahlers, S.M.; Stratman, C. et al. (2014). "Can digital pathology result in cost savings? A financial projection for digital pathology implementation at a large integrated health care organization". Journal of Pathology Informatics 5 (1): 33. doi:10.4103/2153-3539.139714. PMC PMC4168664. PMID 25250191. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4168664.
- ↑ 8.0 8.1 Daniel, C.; García Rojo, M.; Bourquard, K. et al. (2009). "Standards to support information systems integration in anatomic pathology". Archives of Pathology & Laboratory Medicine 133 (11): 1841–9. doi:10.1043/1543-2165-133.11.1841. PMID 19886721.
- ↑ 9.0 9.1 Stathonikos, N.; Veta, M.; Huisman, A.; van Diest, P.J. (2013). "Going fully digital: Perspective of a Dutch academic pathology lab". Journal of Pathology Informatics 4: 15. doi:10.4103/2153-3539.114206. PMC PMC3709427. PMID 23858390. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3709427.
- ↑ 10.0 10.1 Thorstenson, S.; Molin, J.; Lundström, C. (2014). "Implementation of large-scale routine diagnostics using whole slide imaging in Sweden: Digital pathology experiences 2006-2013". Journal of Pathology Informatics 5: 14. doi:10.4103/2153-3539.129452. PMC PMC4023034. PMID 24843825. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4023034.
- ↑ Isaacs, M.; Lennerz, J.K.; Yates, S. et al. (2011). "Implementation of whole slide imaging in surgical pathology: A value added approach". Journal of Pathology Informatics 2: 39. doi:10.4103/2153-3539.84232. PMC PMC3169923. PMID 21969920. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3169923.
- ↑ 12.0 12.1 Kurian, T.; Sharma, C.; Sears, A.; Wilkinson, J. (September 2000). "Front-End and Back-End Integration". Oracle8i Integration Server Overview Release 3 (8.1.7). Oracle Corporation. https://docs.oracle.com/cd/A87860_01/doc/ois.817/a83729/adoisap2.htm. Retrieved 19 April 2016.
- ↑ Papazoglou, M.P.; Georgakopoulos, D. (2003). "Introduction: Service-oriented computing". Communications of the ACM 46 (10): 24–28. doi:10.1145/944217.944233.
- ↑ "HL7 Version 2 Product Suite". HL7 International. http://www.hl7.org/implement/standards/product_brief.cfm?product_id=185. Retrieved 19 April 2016.
- ↑ van de Wetering, R.; Batenburg, R. (2009). "A PACS maturity model: A systematic meta-analytic review on maturation and evolvability of PACS in the hospital enterprise". International Journal of Medical Informatics 78 (2): 127–40. doi:10.1016/j.ijmedinf.2008.06.010. PMID 18752988.
- ↑ Kalinski, T.; Hofmann, H.; Franke, D.S.; Roessner, A. (2002). "Digital imaging and electronic patient records in pathology using an integrated department information system with PACS". Pathology, Research and Practice 198 (10): 679-84. doi:10.1078/0344-0338-00320. PMID 12498223.
- ↑ Krupinski, E.A. (2010). "Optimizing the pathology workstation "cockpit": Challenges and solutions". Journal of Pathology Informatics 1: 19. doi:10.4103/2153-3539.70708. PMC PMC2956171. PMID 21031008. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2956171.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.