Journal:Fueling clinical and translational research in Appalachia: Informatics platform approach
| Full article title | Fueling clinical and translational research in Appalachia: Informatics platform approach |
|---|---|
| Journal | JMIR Medical Informatics |
| Author(s) | Cecchetti, Alfred A.; Bhardwaj, Niharika; Murughiyan, Usha; Kothakapu, Gouthami; Sundaram, Uma |
| Author affiliation(s) | Joan C. Edwards School of Medicine at Marshall University |
| Primary contact | Email: cecchetti at marshall dot edu |
| Year published | 2020 |
| Volume and issue | 8(10) |
| Article # | e17962 |
| DOI | 10.2196/17962 |
| ISSN | 2291-9694 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://medinform.jmir.org/2020/10/e17962/ |
| Download | https://medinform.jmir.org/2020/10/e17962/PDF (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Background: The Appalachian population is distinct, not just culturally and geographically but also in its healthcare needs, facing the most health care disparities in the United States. To meet these unique demands, Appalachian medical centers need an arsenal of analytics and data science tools with the foundation of a centralized data warehouse to transform healthcare data into actionable clinical interventions. However, this is an especially challenging task given the fragmented state of medical data within Appalachia and the need for integration of other types of data such as environmental, social, and economic with medical data.
Objective: This paper aims to present the structure and process of the development of an integrated platform at a midlevel Appalachian academic medical center, along with its initial uses.
Methods: The Appalachian Informatics Platform (AIP) was developed by the Appalachian Clinical and Translational Science Institute’s Division of Clinical Informatics and consists of four major components: a centralized clinical data warehouse, modeling (statistical and machine learning), visualization, and model evaluation. Data from different clinical systems, billing systems, and state- or national-level data sets were integrated into a centralized data warehouse. The platform supports research efforts by enabling curation and analysis of data using the different components, as appropriate.
Results: The AIP is functional and has supported several research efforts since its implementation for a variety of purposes, such as increasing knowledge of the pathophysiology of diseases, risk identification, risk prediction, and healthcare resource utilization research and estimation of the economic impact of diseases.
Conclusions: The platform provides an inexpensive yet seamless way to translate clinical and translational research ideas into clinical applications for regions similar to Appalachia that have limited resources and a largely rural population.
Keywords: Appalachian region, medical informatics, health care disparities, electronic health records, data warehousing, data mining, data visualization, machine learning, data science
Introduction
Background: Unique challenges in Appalachia
Appalachia, with its predominantly rural communities, is known to have one of the worst sets of healthcare outcomes in the United States. This is especially true of southern and central rural Appalachia, which face some of the most severe health disparities in the nation. [1] Over the years, the gap in the overall health between Appalachia and the nation as a whole has continued to grow. [2,3] To close this gap, it is critical to identify the cause of these disparities and direct efforts toward developing necessary interventions to address them.
Such an effort necessitates the adoption of modern technologies such as a centralized research data warehouse to house all data necessary to obtain a comprehensive picture of the health of the Appalachian population before analysis to gain actionable insights can be performed. A centralized data warehouse, once considered strictly a business tool, has evolved into an important instrument for cost containment, tracking of patient outcomes, providing [Clinical decision support system|clinical decision support]] at the point of care, improving prognostic accuracy, and facilitating research. [4] Thus, rural academic medical centers have moved toward implementing data warehouse systems that feed analytical systems for research needs. [5] This entails (1) the integration of data from different types of medical settings (i.e., multi-institutional) such as hospitals, clinics, and specialty centers; (2) linkage of financial data with clinical data, a well-established practice proven to be pivotal to high-quality care and great economic outcomes [6,7]; and (3) integration of other determinants of health such as environmental [8], social [9], and spiritual factors [10] to create longitudinal health records across the care continuum.
However, there are challenges in creating a multi-institutional data warehouse. [11] Electronic health records (EHRs) do not easily interact with one another due to the use of nonstandard terminologies and difficulty in understanding the flow of information. Additionally, significant differences exist between rural and urban health systems. [12-16] Unlike their urban counterparts, healthcare data in Appalachia are typically fragmented, existing in silos within dissimilar databases, registries, data collections, and departmental systems. With innovations in medical technology, the list of data sources continues to grow, producing unprecedented amounts of data from all aspects of care, including diagnosis, medication, procedures, laboratory testing, imaging, and patient self-monitoring. [17-21] To complicate matters, the overall health and health behaviors of Appalachians are strongly affected by Appalachia’s unique culture, geography, and health system issues. [22-24] Consequently, Appalachian academic medical centers face the complex challenge of collecting, organizing, standardizing, and analyzing these enormous quantities of heterogeneous data originating from a wide variety of sources to address the unmet needs of the population they serve.
Why an informatics platform?
Data integration and interoperability have been shown to be key to unlocking these data for data analytics, enabling the development of novel patient management strategies for rural hospitals [25,26] and translational research that leads to new approaches at the bedside for prevention, diagnosis, and treatment of disease, which are essential to improving the health of a population. [27-29] Data analytics, once the domain of the statistician, has now become an equal partner in clinical research and research operations. [30,31] Following the data explosion, data analytics increasingly involves the use of visual analytics tools such as Tableau (Tableau Software Inc.) and Power BI (Microsoft Corp.) to explore data easily and in a self-service fashion and to clearly and effectively communicate complex ideas [32], especially to those members of the medical community who might not have an intimate understanding of the underlying data. Furthermore, machine learning is gaining importance, especially in the area of predictive analytics, to improve the practice of medicine and to infer potentially innovative risk factors. [28,33-35]
However, these applications (e.g., data warehouse, data analytics, statistical analysis, machine learning, visual analytics) are generally uncoordinated without any overarching governance. Thus, we developed an informatics platform—that is, a suite of interconnected, coordinated applications hosted within an operational environment [36]—called the Appalachian Informatics Platform (AIP), in West Virginia (the only state located entirely in Appalachia) that facilitates interoperable access to integrated information, data visualization, and data analytics, thereby functioning as an excellent basis for clinical and translational research to improve health care.
The goal of this study is to describe the structure and process of development of the AIP and demonstrate its value in supporting clinical and translational research.
Methods
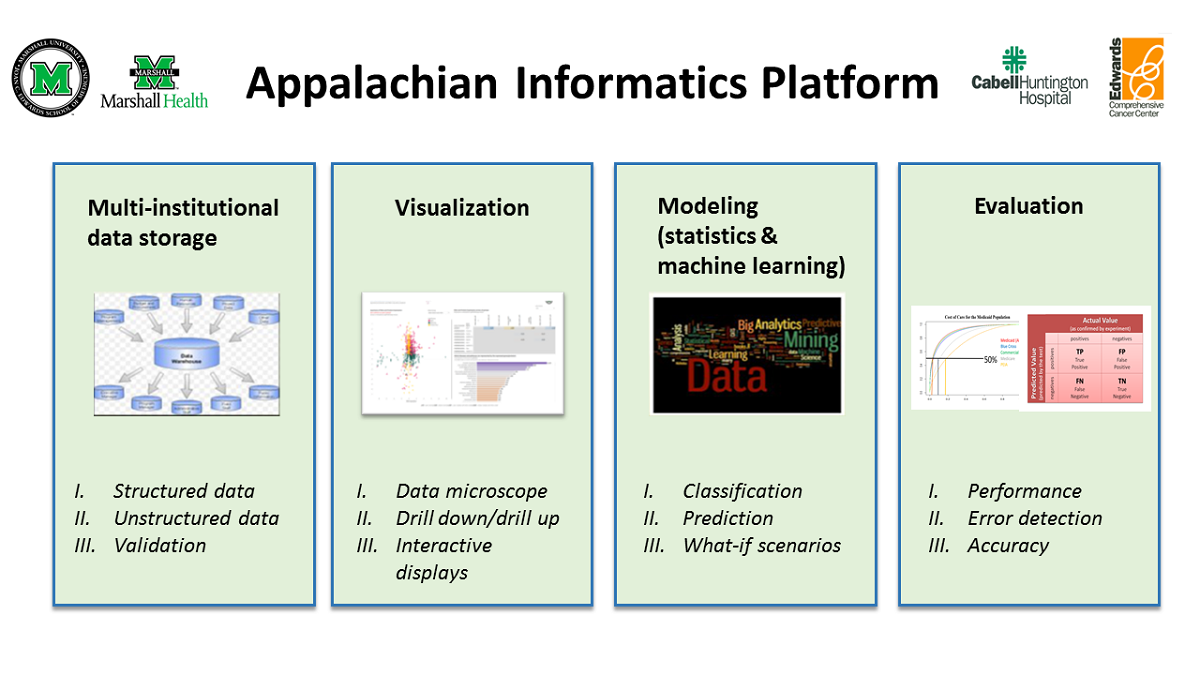
The AIP (Figure 1) is composed of four major components: (1) a multi-institutional data storage or clinical data warehouse (CDW); (2) modeling tools (statistical and machine learning); (3) visualization tools; and (4) evaluation tools. Each of these components is described in detail in separate sections.
The CDW forms an integral part of the AIP. It also contains embedded data analytics (modeling and evaluation) and interactive visualization tools (e.g., Tableau, Power BI). Together, these enable the analysis of Appalachian health information to speed up the transition of translational research ideas into clinical practice.
The CDW serves as a secure source of quality data for descriptive, diagnostic, predictive, and prescriptive analytics for research and operational needs. The visual analytics tools enable an initial exploratory analysis of the processed data and the interactive presentation of analytical findings for further analysis and review. Depending on the use case, data can be analyzed using statistical modeling via external (e.g., SPSS [IBM Corp], Stata [StataCorp]) or integrated (e.g., [[R (programming language)|R] [R Foundation for Statistical Computing], Python [Python Software Foundation] in SQL [Structured Query Language]) applications or machine learning modeling. The performance of the resulting models was evaluated using appropriate metrics. Once trained and evaluated, machine learning models can be deployed and stored in the CDW for future use if needed. Furthermore, the stored machine learning models can be continuously evaluated and improved as more data are generated.
|
The informatics committee governs the access to and utilization of AIP and ensures adherence to security and privacy rules. In addition, team-building activities are also incorporated into our clinical informatics model to foster the development of an effective clinical informatics team.
Multi-institutional data storage: The Appalachian Clinical and Translational Science Institute-Clinical Data Warehouse
The Appalachian Clinical and Translational Science Institute (ACTSI)’s Division of Clinical Informatics solicited buy-in from different entities—namely Cabell-Huntington Hospital (CHH), Edwards Comprehensive Cancer Institute (ECCC), Marshall Health (MH) practice plan, and Marshall University Joan C. Edwards School of Medicine (MU JCESOM)—to build the Appalachian Clinical and Translational Science Institute-Clinical Data Warehouse (ACTSI-CDW) in West Virginia. An agreement was created between these entities that provided access to both financial and clinical data.
The multi-institutional CDW contains more than nine years of billing and clinical data. It comprises relational tables and dimension and fact tables (Online Analytical Processing [OLAP] cube), which enable secure data storage and data access. Designed from the start to facilitate information flow, the CDW can send out a stream of near real-time data that can be used for any authorized research purpose. Documentation includes a data dictionary and flowcharts. Flowcharts follow the patient from admission (or appointment, if outpatient) to discharge (or exit, if outpatient). The data dictionary contains the standardized and source field names, descriptions, and properties, along with the associated metadata for the data contained within the data warehouse. For instance, (1) the entry of a patient into any medical service (admission or appointment) was combined with the single term encounter, and (2) a higher level of precision was introduced by separating patient age into two variables: current age or the age when the procedure was performed.
The CDW process is based on an older data warehouse process developed at the University of Pittsburgh. [37] The process is as follows:
- Data dictionaries are created by recording institutional source field names and field properties and linking them to the standardized CDW names and properties found within the CDW databases. Descriptions of each field (source and CDW) are included.
- Individual institutional flowcharts show the workflow of the data and the location of the people responsible for the quality of the data, which are also used for quality control purposes.
- At present, the CDW contains data from six institutional software packages hosted in various parts of the country (e.g., Cerner data from Kansas City, Missouri; McKesson data from North Druid Hills, Georgia; etc.). The data are exported in a standard format (i.e., ASCII flat file, XML, etc.) and transferred through secure file transfer protocol (e.g., Cerberus [Cerberus, LLC]) to the CDW Development server.
- The data are integrated into the Microsoft SQL databases using Microsoft SQL Server Integration Services (SSIS), a graphical tool that extracts, transforms, and loads (ETL) the data to target schemas that will be used to contain the target data objects: relational tables, dimensions, and cubes. ETL systems enable a smooth migration from one system to another irrespective of the underlying storage system.
- Conformed dimensions were developed, and patient linkages using various methods (e.g., simple heuristics) [38] were also available and made at this time.
- At present, a transactional grain fact table has been developed, but other fact tables will be created as needed.
- The CDW contains internal structured billing and EHR data (i.e., demographics, encounter details, vitals, medications, procedures, diagnoses, orders, immunizations, laboratory and imaging results, date and time, payee, and provider). It also contains unstructured EHR data (e.g., H&P, admission notes, discharge summaries, other clinical notes). These data are received from MH, CHH, and MU JCESOM’s ECCC, as well as from other outside institutions. In addition, non-EHR data are incorporated using REDCap.
- Unstructured data are analyzed using text analytics tools, and classification variables based on text mining are incorporated into the CDW.
- The data structure (OLAP cubes and relational tables), once checked and verified, is transferred from the secure development server to the secure production server for use.
- Various security measures (e.g., IP and password restrictions) are in place to prevent unauthorized use.
- The CDW structure, which stores multi-institutional medical information, can now provide data for both operational and research analytical model development (statistical or machine learning) using very simple de-identified interfaces (e.g., Excel [Microsoft Corp]) or more complex interactive tools (e.g., R, Tableau, Power BI, etc.). Within the CDW, the data can be manipulated, cleaned, and prepared before the analysis as needed.
- Structured and unstructured data currently exist within the CDW. Image and BioSample data will soon be incorporated (like the Pittsburgh model), but the full design has not been finalized yet. An Honest Broker person assumes control of sample shipping and receiving.
- Standard operating procedures (SOPs) have been developed for administrative and technical areas.
- The Health Insurance Portability and Accountability Act (HIPAA) guidelines are followed, and protocol to protect patient information has also been implemented.
The CDW is contained within a Microsoft SQL database that can interact with outside objects using other electronic methods such as SignalR, a software library for Microsoft ASP.NET that allows server code to send asynchronous notifications to client-side web applications, and SqlDependency, an object that represents a query notification dependency between an application and an instance of SQL server. Objects such as these provide the ability for the data warehouse to interact in real time with the outside regional population using the newest technologies such as Microsoft Machine Learning Server with embedded R or Python procedure coding.
Data validation
The information derived from multiple data sources can have inconsistencies and missing values because of their heterogeneous nature, which requires correction. [39-42] Thus, for each research study, clinical and translational researchers using the data warehouse are required to verify a random sample (calculated on the basis of the size of the study population) of all extracted study data are directly verified at the original data source to ensure data accuracy and validity. Identified errors or omissions are transmitted back to the host systems for correction or inclusion.
Augmenting the CDW using REDCap
For certain studies, data available in the CDW may not be precise enough or include variables needed to perform this study. For such studies, data can be augmented using data capture tools. One such tool is the Research Electronic Data Capture, or REDCap, a workflow methodology and software solution designed for the rapid development and deployment of electronic data capture tools to support clinical and translational research [43-45].
Our institution has deployed and maintains 2 REDCap servers: secure (located under institutional firewall) and global (outside the firewall). The secure REDCap system is used for storing data considered protected health information (PHI) under HIPAA. The global system, on the other hand, is used to store de-identified or non-PHI data. These data are then transferred to and stored within the multi-institutional data warehouse. This method of augmenting the information pulled from the existing source systems provides research-grade data from outside sources that are normally not contained within a data warehouse.
Visualization
Visualization of information is an excellent method of providing knowledge that can be easily understood by any member of the health care discipline. Within the informatics platform, Tableau provides interactive drill-down and drill-up capabilities for specific projects.
Tableau is a visual analytics tool that provides an interactive method of exploring multidimensional data, optimized from the data warehouse and OLAP data sources. Tableau, using either indexed relational tables or a data cube, can perform associated operations such as slice, dice, roll-up, and drill-down on the data, providing detailed interactive visual overlays that range from the lowest grain of the data to high-level representations of the data. Tableau charts, graphs, filters, and maps can provide visualization of the various subgroups of interest using a storyboard approach that presents a specific question followed by an interactive dashboard that explores that question in detail. The use of visual elements such as logos, pictograms, icons, or pictures into the dashboards, in association with the subgroups, provides easy-to-reference image aids that provide clarity and understanding of complex information. The data warehouse provides the drill-down, drill-up and slice and dice capability, whereas the hub design connects both financial and clinical data to provide a full picture.
The developed interactive dashboards are securely shared with users within a department or a team, as needed, through the use of Tableau Server. [46]
Modeling (statistics and machine learning)
The modeling component of the informatics platform supports the construction of tailored regional models (statistical or machine learning) to understand and predict disease and other medical events within this region. EHR is primarily a billing system, research only being a secondary function and, thus, is heterogeneous, incomplete, and noisy [25], leading to unrepresentative samples, selection bias, and misclassification. [47] During the modeling process, these issues are eliminated or minimized.
To assist in modeling, software packages such as SPSS and Stata, as well as embedded open-source machine learning programs (e.g., R, Python) are used. This enables faster and easier development of classification, regression, and clustering algorithms for research use. In addition, we utilize products such as Microsoft’s LINQ to electronically gather information and directly incorporate that information into the CDW.
Evaluation
During the modeling process, evaluation of the data set as it relates to the regional population is carried out. Local experts native to this region are asked to evaluate the model from a clinical as well as a financial standpoint. Poverty is endemic within the Appalachian population, and a model that suggests the use of a very expensive medication or procedure over an older but less expensive medication or procedure is unlikely to be used. [48] Thus, the model must take into account whether the patient has the means and access to the recommended medication or procedure. [49] In addition, the willingness of Appalachian medical institutions and health care providers to follow the model’s suggestions must also be evaluated.
Once developed, the models were tuned and tested. Location, time of treatment, outside temperature, and other contributory factors available within the CDW were employed to fine-tune the models, as applicable. The performance of the models was measured using the R programming environment using measures such as area under curve, sensitivity, specificity, F1 score, precision, recall, etc.
Security, privacy, and the informatics committee
Data access and usage are permitted only as described in the mutual agreement between the three institutions and are subject to internal security and privacy rules. All data requests must follow the standard operating procedure built on the basis of mutual multi-institutional agreement. Foremost, the researcher must have appropriate credentials and authorization to be able to request for data. If the researcher is authorized to make requests, he or she must obtain the IRB approval for his or her proposed study and submit the IRB proposal and supporting documentation for review by the informatics committee. The informatics committee, independent of the IRB, reviews all requests for data from the data warehouse to ensure compliance with the agreement. If the research project is approved, the research team designated members are scheduled for the de-identified data extraction process.
Team building
Integral to the informatics platform is team building that builds upon previous work. [37] To facilitate effective team meetings and inter-professional collaboration (local and global) without the need or expense of constant travel, a permanent clinical informatics conference room with a fixed connected computer, an uninterruptable power supply (UPS), a smart board, a camera, and a speaker system, along with a video conferencing system (Zoom) connectivity, was built. This ensures adequate communication among all those involved (i.e., team members, users, leadership, etc.) and access to resources that would otherwise be unavailable.
Results
Since the implementation of the platform, several studies have been conducted. Each study listed below was approved by the informatics committee, and the de-identified data and platform tools were made available securely to the research team.
To evaluate the functionality and value of this platform, we first analyzed the aggregated data of Medicaid-insured patients across different health systems using the interconnected applications within the platform for population health management. Relevant data were extracted from the CDW, followed by exploratory analysis using a Tableau dashboard. Due to the isolated nature of the study population, regional variables such as distance from the CHH and weather conditions (i.e., temperature) were also included. Errors and missing values were identified using the dashboard, and data were subsequently cleaned and prepared. Using these clean data, the regional population was classified into three spend categories: low cost, acute, and persistent subgroups on the basis of the charges accrued. Next, the Charlson Comorbidity Index (CCI) was incorporated into the CDW to predict mortality risk within one year of hospitalization for patients with comorbid conditions within each spend category (Table 1). [50,51]
| ||||||||||||
Of these categories, the persistent group had the largest percentage of patients with a high risk of mortality, followed by acute and low cost after excluding the deceased patients (persistent: 898/1247, 72.01%; acute: 2074/6946, 29.86%; low cost: 5130/102,814, 4.99%). The CCI was not very sensitive in predicting the risk of mortality but was very specific and accurate (sensitivity: 896/1512, 59.26%; specificity: 102,905/111,007, 92.7%; accuracy: 103,801/112,519, 92.25%). The effect of distance and weather on the CCI needs further investigation that is being conducted. Adjustments are being made to this standard national index to incorporate other Appalachian characteristics that could improve the sensitivity of this risk scoring system.
As such, the platform has been utilized for a variety of purposes such as increasing knowledge of the pathophysiology of diseases, risk identification, risk prediction, health care resource utilization research, and estimation of the economic impact of diseases to enable data-driven clinical decisions, leading to improved clinical outcomes. Table 2 contains a list of studies conducted so far.
|
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, grammar, and punctuation. In some cases important information was missing from the references, and that information was added.