Journal:Support patient search on pathology reports with interactive online learning based data extraction
| Full article title | Support patient search on pathology reports with interactive online learning based data extraction |
|---|---|
| Journal | Journal of Pathology Informatics |
| Author(s) | Zheng, Shuai; Lu, James J.; Appin, Christina; Brat, Daniel; Wang, Fusheng |
| Author affiliation(s) | Emory University, Stony Brook University |
| Primary contact | Email: N/A |
| Year published | 2015 |
| Volume and issue | 6 |
| Page(s) | 51 |
| DOI | 10.4103/2153-3539.166012 |
| ISSN | 2153-3539 |
| Distribution license | Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported |
| Website | http://www.jpathinformatics.org |
| Download | http://www.jpathinformatics.org/temp/JPatholInform6151-4749188_131131.pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background: Structural reporting enables semantic understanding and prompt retrieval of clinical findings about patients. While synoptic pathology reporting provides templates for data entries, information in pathology reports remains primarily in narrative free text form. Extracting data of interest from narrative pathology reports could significantly improve the representation of the information and enable complex structured queries. However, manual extraction is tedious and error-prone, and automated tools are often constructed with a fixed training dataset and not easily adaptable. Our goal is to extract data from pathology reports to support advanced patient search with a highly adaptable semi-automated data extraction system, which can adjust and self-improve by learning from a user's interaction with minimal human effort.
Methods: We have developed an online machine learning based information extraction system called IDEAL-X. With its graphical user interface, the system's data extraction engine automatically annotates values for users to review upon loading each report text. The system analyzes users' corrections regarding these annotations with online machine learning, and incrementally enhances and refines the learning model as reports are processed. The system also takes advantage of customized controlled vocabularies, which can be adaptively refined during the online learning process to further assist the data extraction. As the accuracy of automatic annotation improves overtime, the effort of human annotation is gradually reduced. After all reports are processed, a built-in query engine can be applied to conveniently define queries based on extracted structured data.
Results: We have evaluated the system with a dataset of anatomic pathology reports from 50 patients. Extracted data elements include demographical data, diagnosis, genetic marker, and procedure. The system achieves F-1 scores of around 95% for the majority of tests.
Conclusions: Extracting data from pathology reports could enable more accurate knowledge to support biomedical research and clinical diagnosis. IDEAL-X provides a bridge that takes advantage of online machine learning based data extraction and the knowledge from human's feedback. By combining iterative online learning and adaptive controlled vocabularies, IDEAL-X can deliver highly adaptive and accurate data extraction to support patient search.
Keywords: Controlled vocabularies, data extraction, online machine learning, pathology reports, patient search
Introduction
Pathology reports contain valuable research information embedded in narrative free text. The same information in structured format can be used to support clinical findings, decision making and biomedical research. Synoptic reporting[1][2][3] has become a powerful tool for providing summarized findings through predefined data element templates such as CAP Cancer Protocols.[4] Meanwhile, standard groups such as IHE are proposing structured reporting standards such as Anatomic Pathology Structured Reports[5] in Health Level Seven. While there is a major trend for structured reporting, the vast amount of pathology reports remain unstructured in legacy systems. And standardization efforts only capture major data elements, leaving a substantial amount of valuable information in free text that is difficult to process and search.
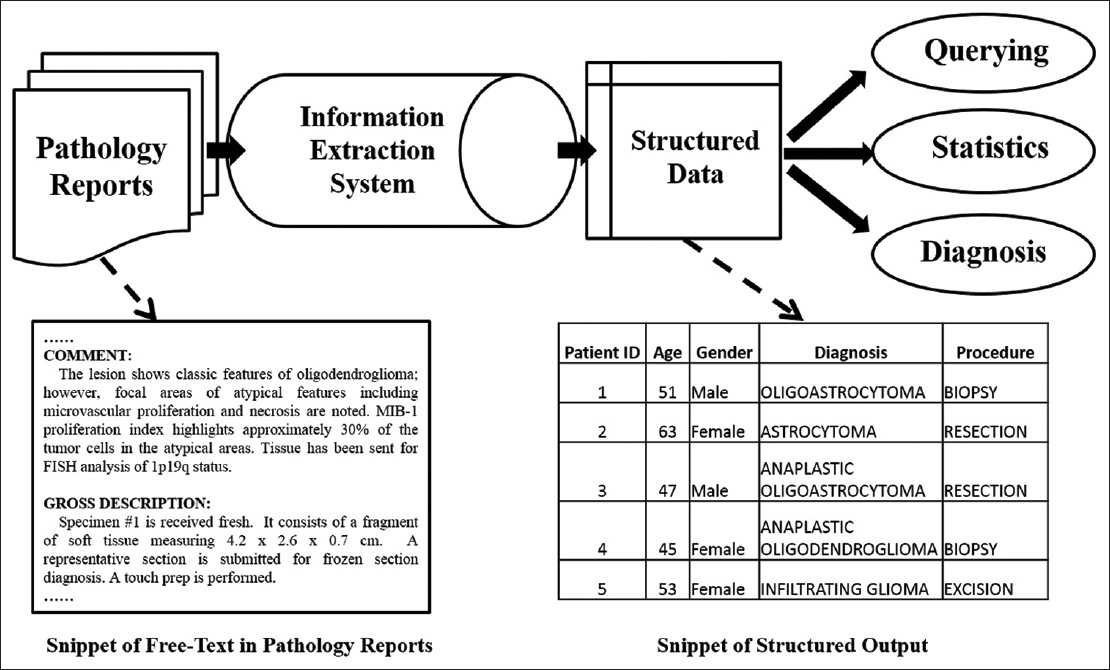
Information extraction is a technique that can generate structured representation of important information from pathology reports. The transformed data may be used to search easily for patient groups with certain traits as in, for example, find all patients with an age above 40 years old and that have a diagnosis glioma. Figure 1 shows a typical workflow of data extraction from pathology reports.
Figure 1. Common pipeline of processing free text medical report
Previous work on data extraction from pathology reports addresses various tasks and different research problems. caTIES supports coding for surgical pathology reports.[6] A regular expression is used to mine specimens and related information in[7], MedTAS/P extracts and represents cancer diseases from pathology reports with the hierarchical model.[8] Lupus represents extracted information with Semantic Web techniques.[9] NegEx is adopted to detect negation for annotating surgical pathology report.[10] These systems either employ rules engineered to specific topics and domains or they use statistical models learned in batch from manually annotated training data. The first approach lacks generalizability; new rules need to be designed and developed for each domain. The second approach based on machine learning is more flexible. But obtaining accurate training data can be costly and time-consuming.
We present a system, IDEAL-X, which combines online machine learning and customizable vocabularies to provide a generic, easy-to-use solution for clinical information extraction. Online machine learning[11][12][13] takes an iterative learning approach through interactive human intervention, the data extraction engine of IDEAL-X automatically predicts answers to annotate reports, gradually learns from human's feedback, and incrementally improves its accuracy. Compared to traditional batch training based algorithm, which requires pretraining with a reasonably large dataset, online learning based algorithms can significantly reduce human effort on labeling training data and provide the possibility of updating the learning models dynamically to fit a continually changing data environment. To enhance its performance, IDEAL-X supports adaptive vocabulary to support data extraction. A user can customize a controlled vocabulary, which could be continuously adjusted during online learning process. Once structured data elements are extracted, a query interface is provided to support patient search with filtering conditions on data elements.
References
- ↑ Srigley, J.R.; McGowan, T.; Maclean, A.; Raby, M.; Ross, J.; Kramer, S.; Sawka, C. (2009). "Standardized synoptic cancer pathology reporting: A population-based approach". Journal of Surgical Oncology 99 (8): 517–524. doi:10.1002/jso.21282. PMID 19466743.
- ↑ Gill, A.J.; Johns, A.L.; Eckstein, R.; Samra, J.S.; Kaufman, A.; Chang, D.K.; Merrett N.D.; Cosman, P.H.; Smith, R.C.; Biankin, A.V.; Kench, J.G.; New South Wales Pancreatic Cancer Network (2009). "Synoptic reporting improves histopathological assessment of pancreatic resection specimens". Pathology 41 (2): 161–167. PMID 19320058.
- ↑ Leslie, K.O.; Rosai, J. (1994). "Standardization of the surgical pathology report: Formats, templates, and synoptic reports". Seminars in Diagnostic Pathology 11 (4): 253–7. PMID 7878300.
- ↑ "Cancer Protocols". CAP.org. College of American Pathologists. 2015. http://www.cap.org/web/oracle/webcenter/portalapp/pagehierarchy/cancer_protocols.jspx. Retrieved 12 July 2015.
- ↑ "Anatomic Pathology Structured Reports". IHE Wiki. Integrating the Healthcare Enterprise. 30 June 2010. http://wiki.ihe.net/index.php?title=Anatomic_Pathology_Structured_Reports. Retrieved 12 July 2015.
- ↑ Crowley, R.S.; Castine, M.; Mitchell, K.; Chavan, G.; McSherry, T.; Feldman, M. (2010). "caTIES: A grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research". Journal of the American Medical Informatics Association 17 (3): 253–64. doi:10.1136/jamia.2009.002295. PMC PMC2995710. PMID 20442142. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2995710.
- ↑ Schadow, G.; McDonald, C.J. (2003). "Extracting structured information from free text pathology reports". AMIA Annual Symposium Proceedings 2003: 584–8. PMC PMC1480213. PMID 14728240. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1480213.
- ↑ Coden, A.; Savova, G.; Sominsky, I.; Tanenblatt, M.; Masanz, J.; Schuler, K.; Cooper, J.; Guan, W.; de Groen, P.C. (2009). "Automatically extracting cancer disease characteristics from pathology reports into a disease knowledge representation model". Journal of Biomedical Informatics 42 (5): 937–49. doi:10.1016/j.jbi.2008.12.005. PMID 19135551.
- ↑ Schlangen, David; Stede, Manfred; Bontas, Elena Paslaru (2004). "Feeding Owl: Extracting and Representing the Content of Pathology Reports". NLPXML '04 Proceeedings of the Workshop on NLP and XML (NLPXML-2004): RDF/RDFS and OWL in Language Technology: 43–50. http://pub.uni-bielefeld.de/publication/1992186.
- ↑ Mitchell, K.J.; Becich, M.J.; Berman, J.J.; Chapman, W.W.; Gilbertson, J.; Gupta, D.; Harrison, J.; Legowski, E.; Crowley, R.S. (2004). "Implementation and evaluation of a negation tagger in a pipeline-based system for information extract from pathology reports". Studies in Health Technology and Informatics 107 (Pt 1): 663-7. doi:10.3233/978-1-60750-949-3-663. PMID 15360896.
- ↑ Smale, S.; Yao, Y. (2006). "Online learning algorithms". Foundations of Computational Mathematics 6 (2): 145–170. doi:10.1007/s10208-004-0160-z.
- ↑ Shalev-Shwartz, S. (2012). "Online learning and online convex optimization". Foundations and Trends in Machine Learning 4 (2): 107–194. doi:10.1561/2200000018.
- ↑ Shalev-Shwartz, S. (July 2007). "Online Learning: Theory, Algorithms, and Applications" (PDF). University of Chicago. http://ttic.uchicago.edu/~shai/papers/ShalevThesis07.pdf. Retrieved 12 July 2015.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In several cases the PubMed ID was missing and was added to make the reference more useful.