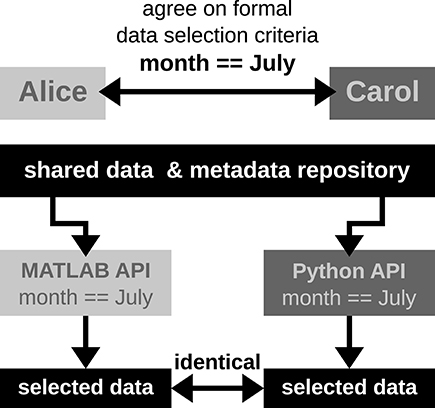
Featured article of the week: June 18–24:"Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory"
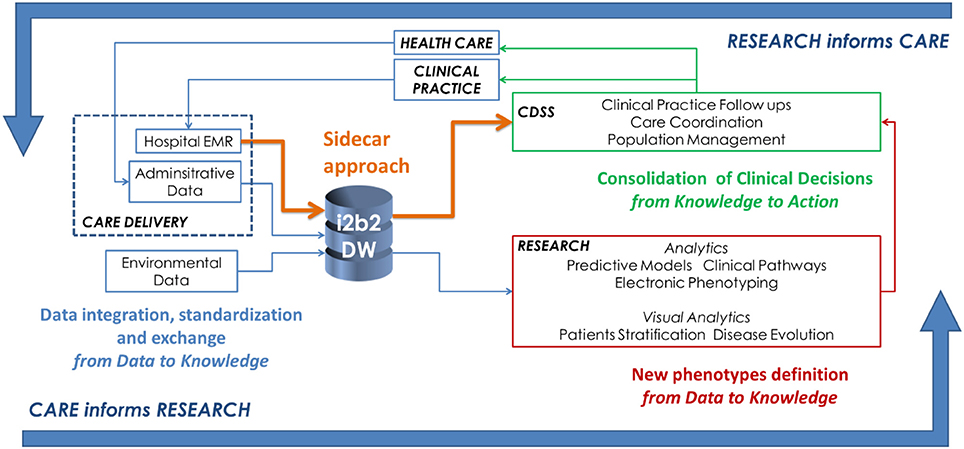
"Big data as a driver for clinical decision support systems: A learning health systems perspective"
Big data technologies are nowadays providing health care with powerful instruments to gather and analyze large volumes of heterogeneous data collected for different purposes, including clinical care, administration, and research. This makes possible to design IT infrastructures that favor the implementation of the so-called “Learning Healthcare System Cycle,” where healthcare practice and research are part of a unique and synergistic process. In this paper we highlight how "big-data-enabled” integrated data collections may support clinical decision-making together with biomedical research. Two effective implementations are reported, concerning decision support in diabetes and in inherited arrhythmogenic diseases. (Full article...)
Featured article of the week: June 11–17:
"Implementation and use of cloud-based electronic lab notebook in a bioprocess engineering teaching laboratory"
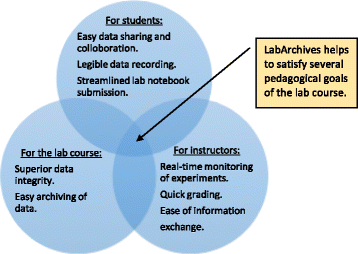
Electronic laboratory notebooks (ELNs) are better equipped than paper laboratory notebooks (PLNs) to handle present-day life science and engineering experiments that generate large data sets and require high levels of data integrity. But limited training and a lack of workforce with ELN knowledge have restricted the use of ELN in academic and industry research laboratories, which still rely on cumbersome PLNs for record keeping. We used LabArchives, a cloud-based ELN in our bioprocess engineering lab course to train students in electronic record keeping, good documentation practices (GDPs), and data integrity.
Implementation of ELN in the bioprocess engineering lab course, an analysis of user experiences, and our development actions to improve ELN training are presented here. ELN improved pedagogy and learning outcomes of the lab course through streamlined workflow, quick data recording and archiving, and enhanced data sharing and collaboration. It also enabled superior data integrity, simplified information exchange, and allowed real-time and remote monitoring of experiments. (Full article...)
|
Featured article of the week: June 4–10:
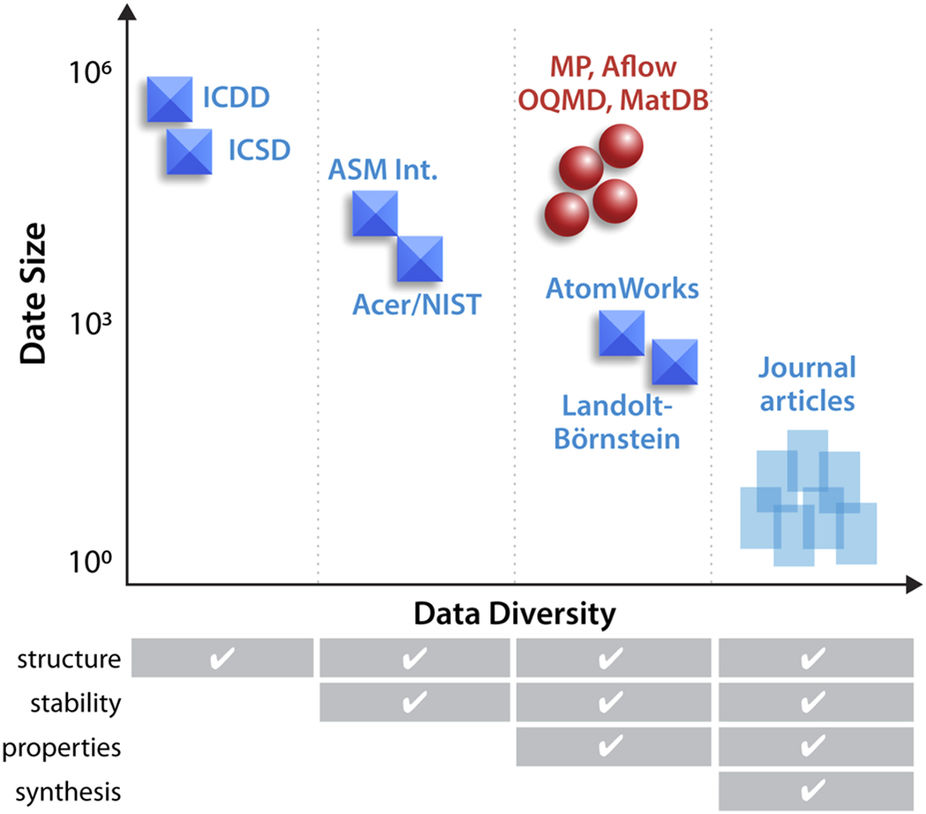
"An open experimental database for exploring inorganic materials"
The use of advanced machine learning algorithms in experimental materials science is limited by the lack of sufficiently large and diverse datasets amenable to data mining. If publicly open, such data resources would also enable materials research by scientists without access to expensive experimental equipment. Here, we report on our progress towards a publicly open High Throughput Experimental Materials (HTEM) Database (htem.nrel.gov). This database currently contains 140,000 sample entries, characterized by structural (100,000), synthetic (80,000), chemical (70,000), and optoelectronic (50,000) properties of inorganic thin film materials, grouped in >4,000 sample entries across >100 materials systems; more than a half of these data are publicly available. This article shows how the HTEM database may enable scientists to explore materials by browsing web-based user interface and an application programming interface. This paper also describes a HTE approach to generating materials data and discusses the laboratory information management system (LIMS) that underpins the HTEM database. Finally, this manuscript illustrates how advanced machine learning algorithms can be adopted to materials science problems using this open data resource. (Full article...)
|
Featured article of the week: May 28–June 3:
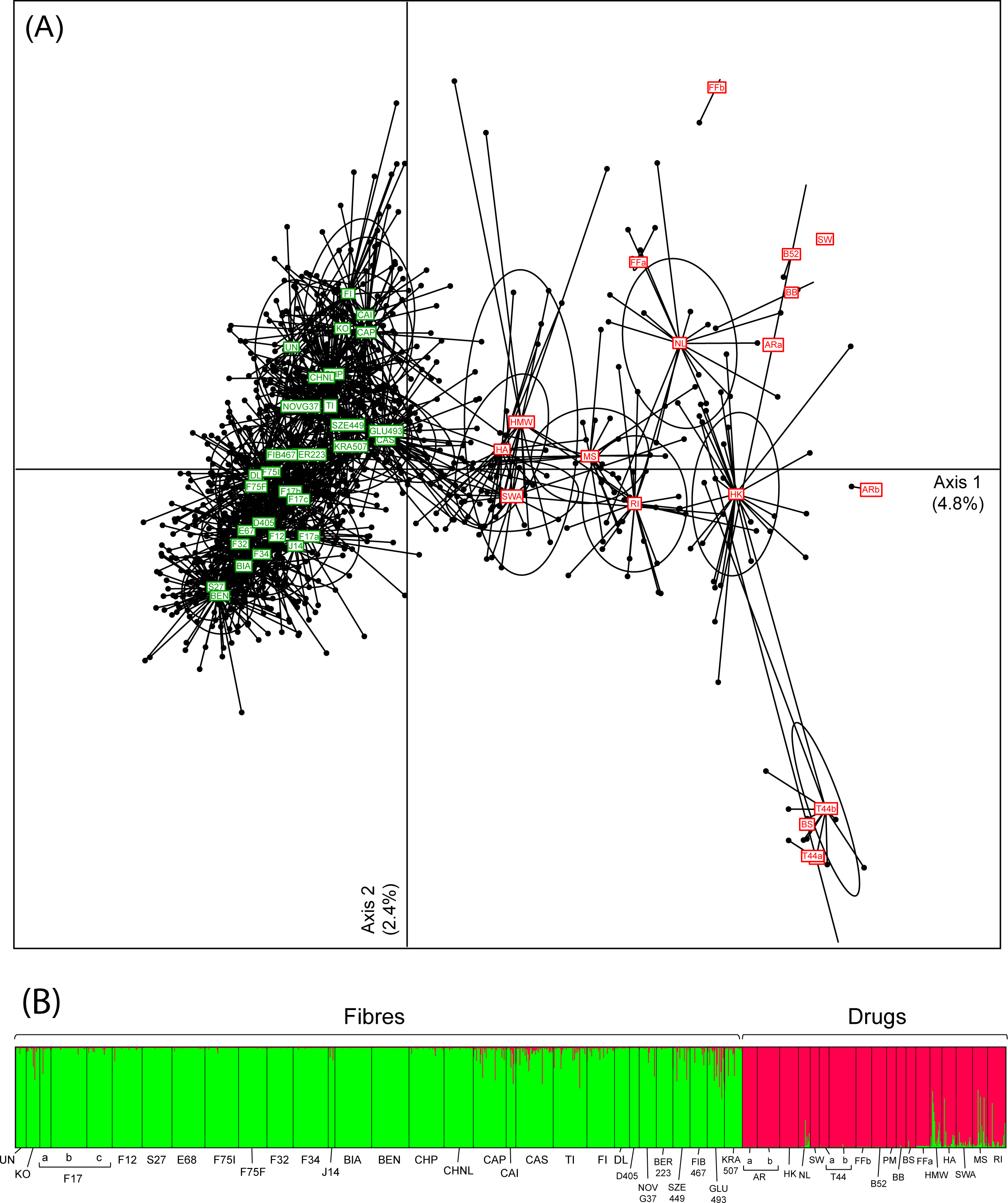
"Broad-scale genetic diversity of Cannabis for forensic applications"
Cannabis (hemp and marijuana) is an iconic yet controversial crop. On the one hand, it represents a growing market for pharmaceutical and agricultural sectors. On the other hand, plants synthesizing the psychoactive THC produce the most widespread illicit drug in the world. Yet, the difficulty to reliably distinguish between Cannabis varieties based on morphological or biochemical criteria impedes the development of promising industrial programs and hinders the fight against narcotrafficking. Genetics offers an appropriate alternative to characterize drug vs. non-drug Cannabis. However, forensic applications require rapid and affordable genotyping of informative and reliable molecular markers for which a broad-scale reference database, representing both intra- and inter-variety variation, is available. Here we provide such a resource for Cannabis, by genotyping 13 microsatellite loci (STRs) in 1,324 samples selected specifically for fiber (24 hemp varieties) and drug (15 marijuana varieties) production. We showed that these loci are sufficient to capture most of the genome-wide diversity patterns recently revealed by next-generation sequencing (NGS) data. (Full article...)
|
Featured article of the week: May 21–27:
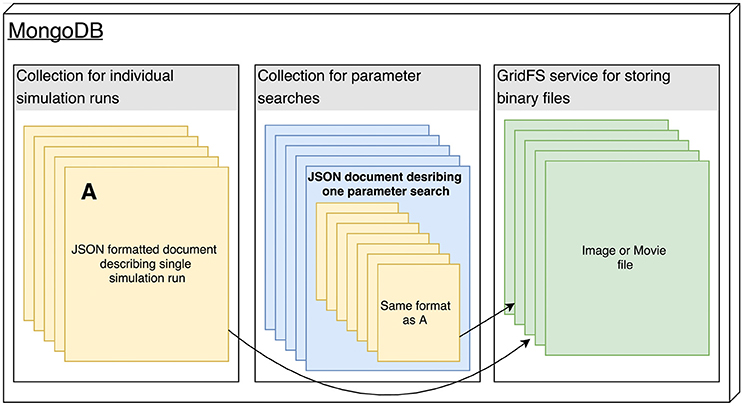
"Arkheia: Data management and communication for open computational neuroscience"
Two trends have been unfolding in computational neuroscience during the last decade. First, focus has shifted to increasingly complex and heterogeneous neural network models, with a concomitant increase in the level of collaboration within the field (whether direct or in the form of building on top of existing tools and results). Second, general trends in science have shifted toward more open communication, both internally, with other potential scientific collaborators, and externally, with the wider public. This multi-faceted development toward more integrative approaches and more intense communication within and outside of the field poses major new challenges for modelers, as currently there is a severe lack of tools to help with automatic communication and sharing of all aspects of a simulation workflow to the rest of the community. To address this important gap in the current computational modeling software infrastructure, here we introduce Arkheia, a web-based open science platform for computational models in systems neuroscience. It provides an automatic, interactive, graphical presentation of simulation results, experimental protocols, and interactive exploration of parameter searches in a browser-based application. (Full article...)
|
Featured article of the week: May 14–20:
"Developing a bioinformatics program and supporting infrastructure in a biomedical library"
Over the last couple decades, the field of bioinformatics has helped spur medical discoveries that offer a better understanding of the genetic basis of disease, which in turn improve public health and save lives. Concomitantly, support requirements for molecular biology researchers have grown in scope and complexity, incorporating specialized resources, technologies, and techniques.
To address this specific need among National Institutes of Health (NIH) intramural researchers, the NIH Library hired an expert bioinformatics trainer and consultant with a PhD in biochemistry to implement a bioinformatics support program. This study traces the program from its inception in 2009 to its present form. Discussion involves the particular skills of program staff, development of content, collection of resources, associated technology, assessment, and the impact of the program on the NIH community. (Full article...)
|
Featured article of the week: May 7–13:
"Big data and public health systems: Issues and opportunities"
In recent years, the need for changing the current model of European public health systems has been repeatedly addressed, in order to ensure their sustainability. Following this line, information technology (IT) has always been referred to as one of the key instruments for enhancing the information management processes of healthcare organizations, thus contributing to the improvement and evolution of health systems. On the IT field, big data solutions are expected to play a main role, since they are designed for handling huge amounts of information in a fast and efficient way, allowing users to make important decisions quickly. This article reviews the main features of the European public health system model and the corresponding healthcare and management-related information systems, the challenges that these health systems are currently facing, and the possible contributions of big data solutions to this field. To that end, the authors share their professional experience on the Spanish public health system and review the existing literature related to this topic. (Full article...)
|
Featured article of the week: April 30–May 6:
"Generating big data sets from knowledge-based decision support systems to pursue value-based healthcare"
Talking about big data in healthcare we usually refer to how to use data collected from current electronic medical records, either structured or unstructured, to answer clinically relevant questions. This operation is typically carried out by means of analytics tools (e.g., machine learning) or by extracting relevant data from patient summaries through natural language processing techniques. From other perspectives of research in medical informatics, powerful initiatives have emerged to help physicians make decisions, in both diagnostics and therapeutics, built from existing medical evidence (i.e., knowledge-based decision support systems). Many of the problems these tools have shown, when used in real clinical settings, are related to their implementation and deployment, more than failing in their support; however, technology is slowly overcoming interoperability and integration issues. (Full article...)
|
Featured article of the week: April 23–29:
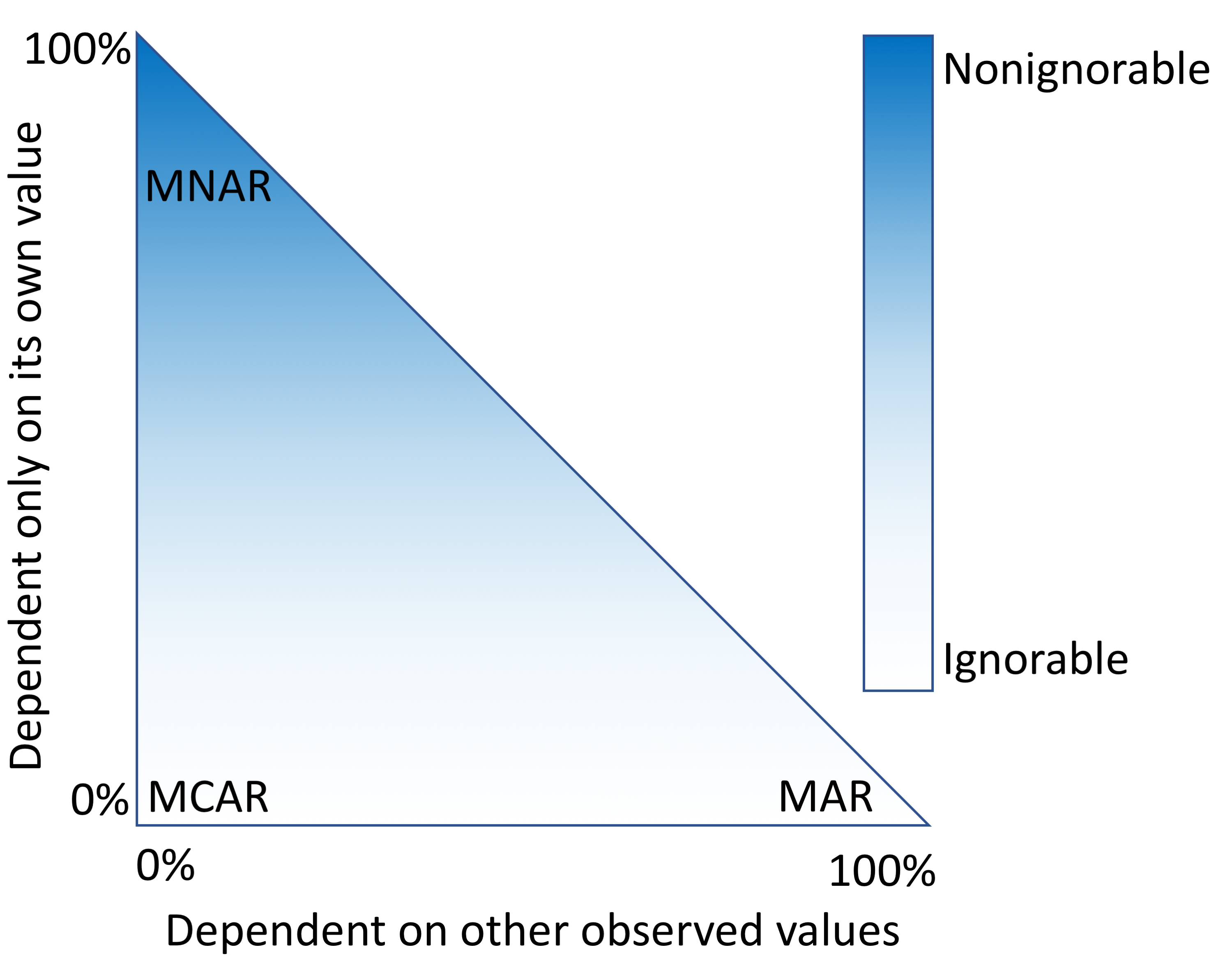
"Characterizing and managing missing structured data in electronic health records: Data analysis"
Missing data is a challenge for all studies; however, this is especially true for electronic health record (EHR)-based analyses. Failure to appropriately consider missing data can lead to biased results. While there has been extensive theoretical work on imputation, and many sophisticated methods are now available, it remains quite challenging for researchers to implement these methods appropriately. Here, we provide detailed procedures for when and how to conduct imputation of EHR laboratory results.
The objective of this study was to demonstrate how the mechanism of "missingness" can be assessed, evaluate the performance of a variety of imputation methods, and describe some of the most frequent problems that can be encountered. (Full article...)
|
Featured article of the week: April 16–22:
"Closha: Bioinformatics workflow system for the analysis of massive sequencing data"
While next-generation sequencing (NGS) costs have fallen in recent years, the cost and complexity of computation remain substantial obstacles to the use of NGS in bio-medical care and genomic research. The rapidly increasing amounts of data available from the new high-throughput methods have made data processing infeasible without automated pipelines. The integration of data and analytic resources into workflow systems provides a solution to the problem by simplifying the task of data analysis.
To address this challenge, we developed a cloud-based workflow management system, Closha, to provide fast and cost-effective analysis of massive genomic data. We implemented complex workflows making optimal use of high-performance computing clusters. Closha allows users to create multi-step analyses using drag-and-drop functionality and to modify the parameters of pipeline tools. (Full article...)
|
Featured article of the week: April 9–15:
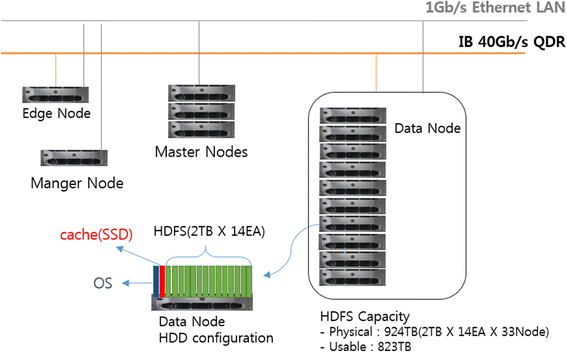
"Big data management for cloud-enabled geological information services"
Cloud computing as a powerful technology of performing massive-scale and complex computing plays an important role in implementing geological information services. In the era of big data, data are being collected at an unprecedented scale. Therefore, to ensure successful data processing and analysis in cloud-enabled geological information services (CEGIS), we must address the challenging and time-demanding task of big data processing. This review starts by elaborating the system architecture and the requirements for big data management. This is followed by the analysis of the application requirements and technical challenges of big data management for CEGIS in China. This review also presents the application development opportunities and technical trends of big data management in CEGIS, including collection and preprocessing, storage and management, analysis and mining, parallel computing-based cloud platforms, and technology applications. (Full article...)
|
Featured article of the week: April 2–8:
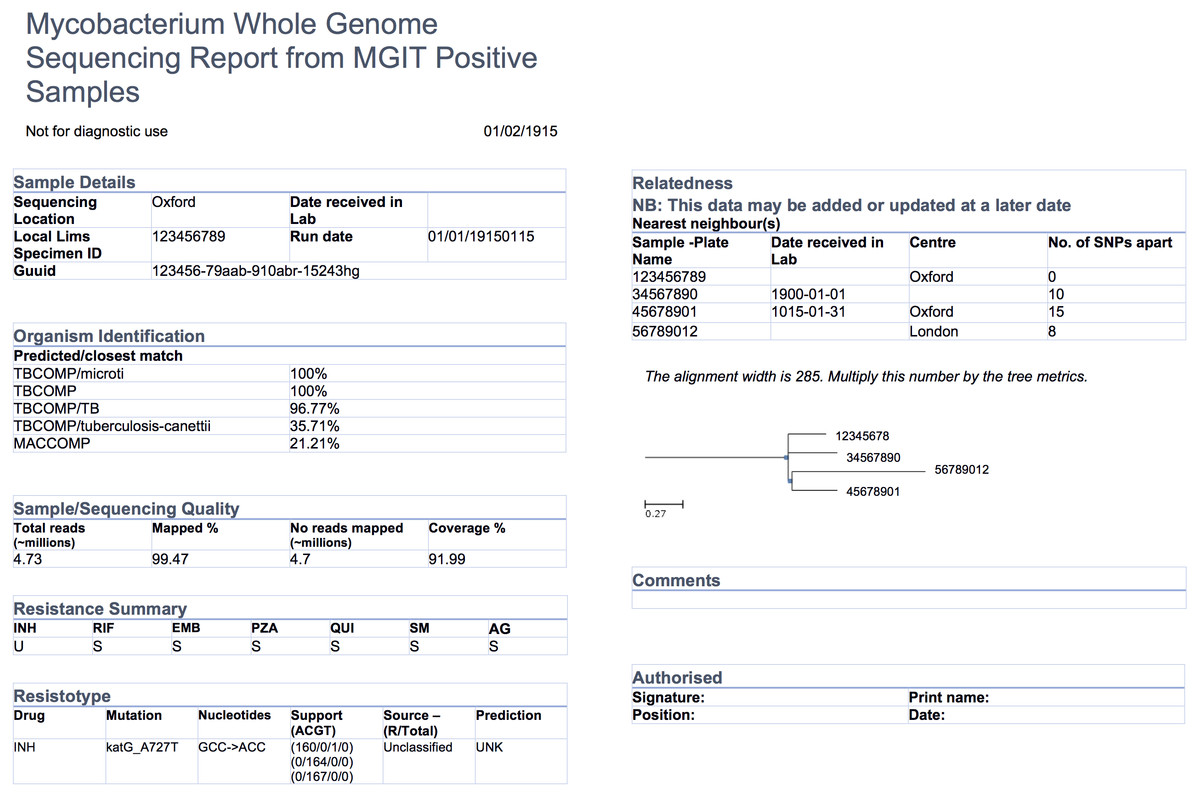
"Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory"
Microbial genome sequencing is now being routinely used in many clinical and public health laboratories. Understanding how to report complex genomic test results to stakeholders who may have varying familiarity with genomics — including clinicians, laboratorians, epidemiologists, and researchers — is critical to the successful and sustainable implementation of this new technology; however, there are no evidence-based guidelines for designing such a report in the pathogen genomics domain. Here, we describe an iterative, human-centered approach to creating a report template for communicating tuberculosis (TB) genomic test results. (Full article...)
|
Featured article of the week: March 26–April 1:
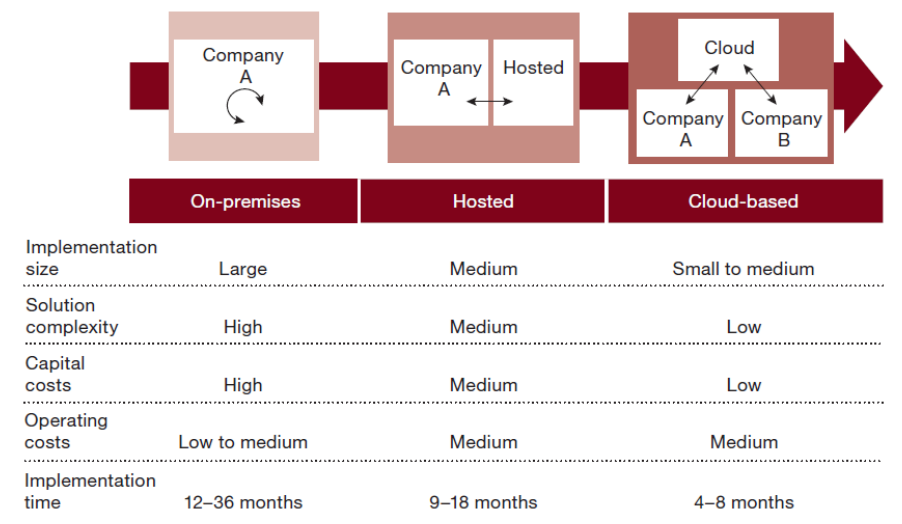
"Moving ERP systems to the cloud: Data security issues"
This paper brings to light data security issues and concerns for organizations by moving their enterprise resource planning (ERP) systems to the cloud. Cloud computing has become the new trend of how organizations conduct business and has enabled them to innovate and compete in a dynamic environment through new and innovative business models. The growing popularity and success of the cloud has led to the emergence of cloud-based software as a service (SaaS) ERP systems, a new alternative approach to traditional on-premise ERP systems. Cloud-based ERP has a myriad of benefits for organizations. However, infrastructure engineers need to address data security issues before moving their enterprise applications to the cloud. Cloud-based ERP raises specific concerns about the confidentiality and integrity of the data stored in the cloud. Such concerns that affect the adoption of cloud-based ERP are based on the size of the organization. (Full article...)
|
Featured article of the week: March 19–25:
"Method-centered digital communities on protocols.io for fast-paced scientific innovation"
The internet has enabled online social interaction for scientists beyond physical meetings and conferences. Yet despite these innovations in communication, dissemination of methods is often relegated to just academic publishing. Further, these methods remain static, with subsequent advances published elsewhere and unlinked. For communities undergoing fast-paced innovation, researchers need new capabilities to share, obtain feedback, and publish methods at the forefront of scientific development. For example, a renaissance in virology is now underway given the new metagenomic methods to sequence viral DNA directly from an environment. Metagenomics makes it possible to “see” natural viral communities that could not be previously studied through culturing methods. Yet, the knowledge of specialized techniques for the production and analysis of viral metagenomes remains in a subset of labs. (Full article...)
|
Featured article of the week: March 12–18:
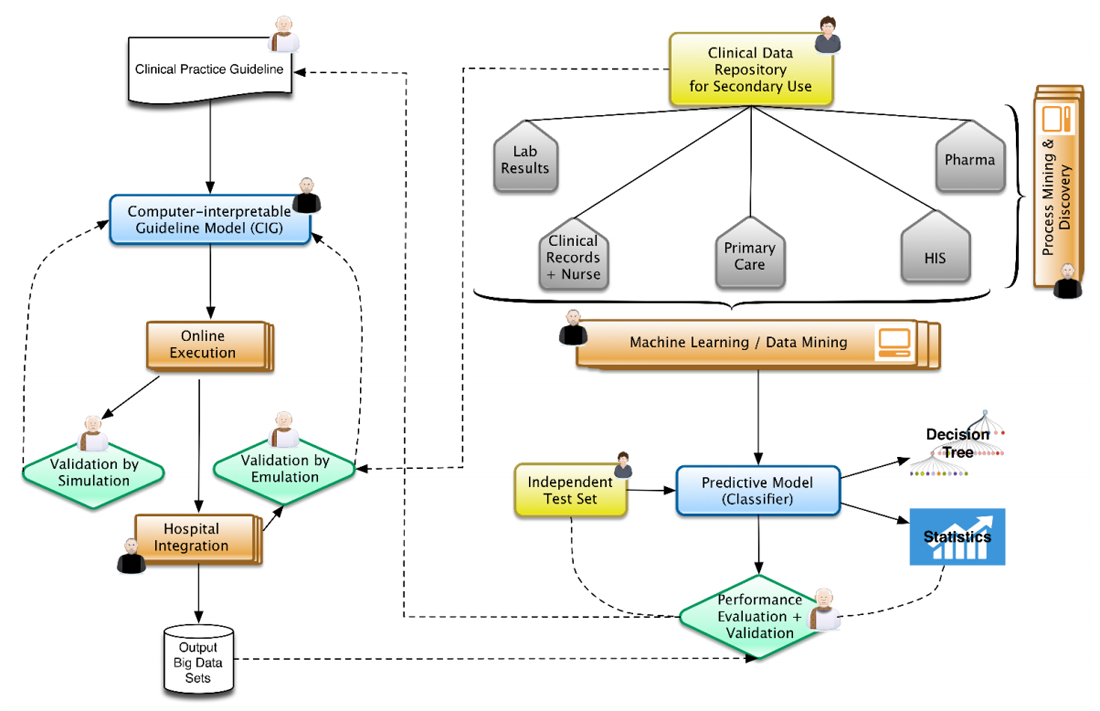
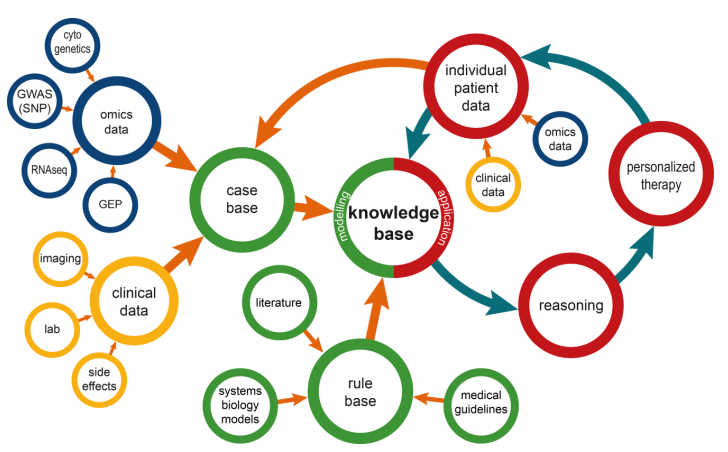
"Information management for enabling systems medicine"
Systems medicine is a data-oriented approach in research and clinical practice to support the study and treatment of complex diseases. It relies on well-defined information management processes providing comprehensive and up-to-date information as the basis for electronic decision support. The authors suggest a three-layer information technology (IT) architecture for systems medicine and a cyclic data management approach, including a knowledge base that is dynamically updated by extract, transform, and load (ETL) procedures. Decision support is suggested as case-based and rule-based components. Results are presented via a user interface to acknowledging clinical requirements in terms of time and complexity. The systems medicine application was implemented as a prototype. (Full article...)
|
Featured article of the week: March 5–11:
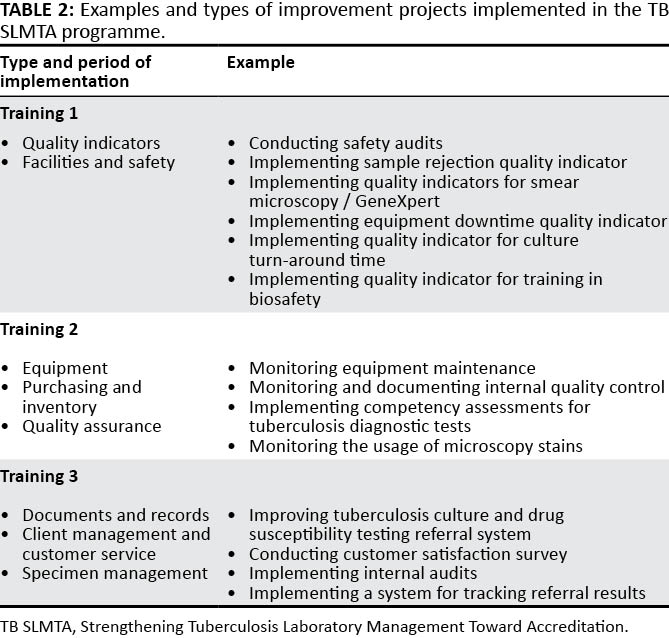
"Developing a customized approach for strengthening tuberculosis laboratory quality management systems toward accreditation"
Quality-assured tuberculosis laboratory services are critical to achieve global and national goals for tuberculosis prevention and care. Implementation of a quality management system (QMS) in laboratories leads to improved quality of diagnostic tests and better patient care. The Strengthening Laboratory Management Toward Accreditation (SLMTA) program has led to measurable improvements in the QMS of clinical laboratories. However, progress in tuberculosis laboratories has been slower, which may be attributed to the need for a structured tuberculosis-specific approach to implementing QMS. We describe the development and early implementation of the Strengthening Tuberculosis Laboratory Management Toward Accreditation (TB SLMTA) program. The TB SLMTA curriculum was developed by customizing the SLMTA curriculum to include specific tools, job aids, and supplementary materials specific to the tuberculosis laboratory. (Full article...)
|
Featured article of the week: February 26–March 4:
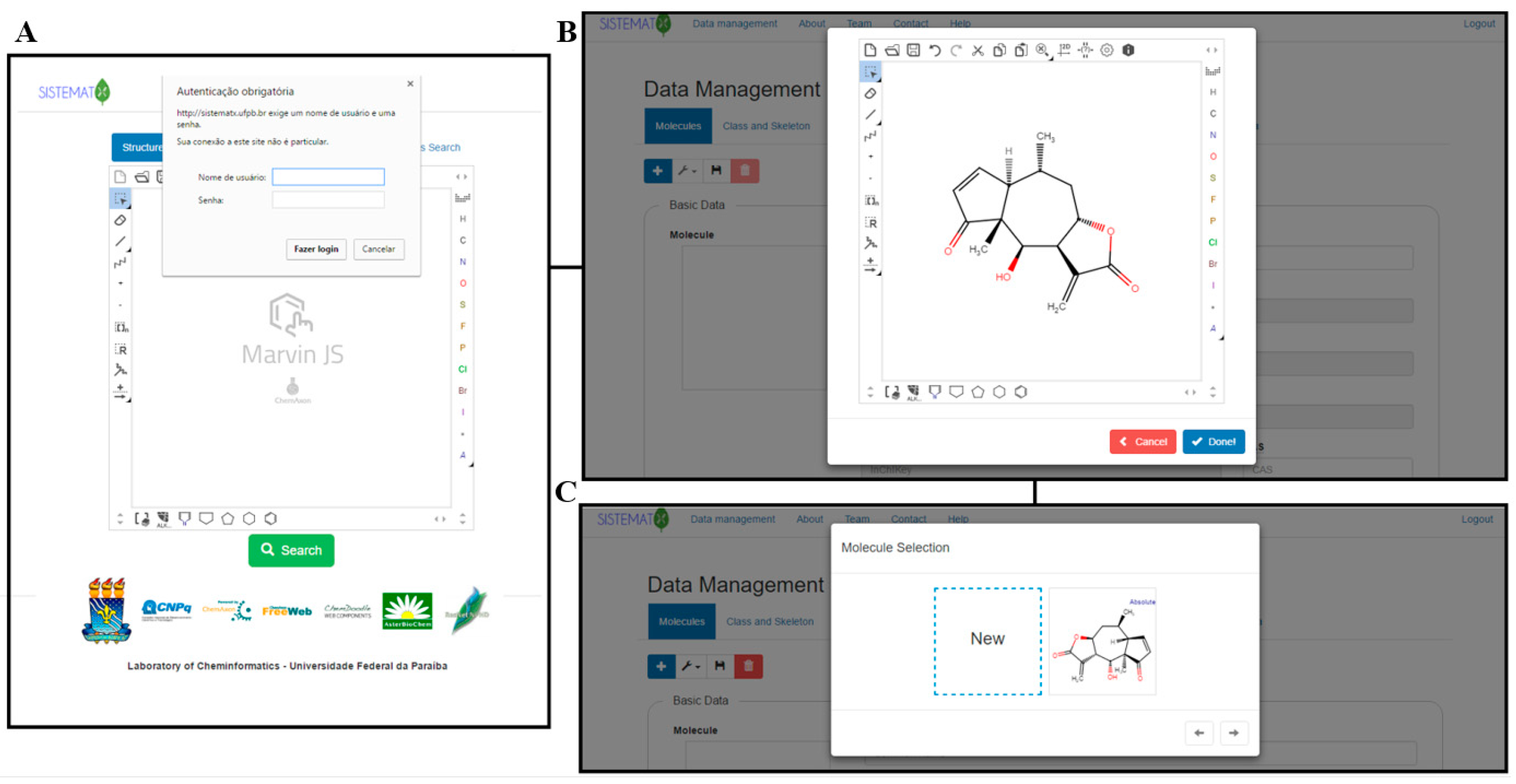
"SistematX, an online web-based cheminformatics tool for data management of secondary metabolites"
The traditional work of a natural products researcher consists in large part of time-consuming experimental work, collecting biota to prepare, extracts to analyze, and innovative metabolites to identify. However, along this long scientific path, much information is lost or restricted to a specific niche. The large amounts of data already produced and the science of metabolomics reveal new questions: Are these compounds known or new? How fast can this information be obtained? To answer these and other relevant questions, an appropriate procedure to correctly store information on the data retrieved from the discovered metabolites is necessary. The SistematX (http://sistematx.ufpb.br) interface is implemented considering the following aspects: (a) the ability to search by structure, SMILES (Simplified Molecular-Input Line-Entry System) code, compound name, and species; (b) the ability to save chemical structures found by searching; (c) the ability to display compound data results, including important characteristics for natural products chemistry; and (d) the user's ability to find specific information for taxonomic rank (from family to species) of the plant from which the compound was isolated, the searched-for molecule, and the bibliographic reference and Global Positioning System (GPS) coordinates. (Full article...)
|
Featured article of the week: February 19–25:
"Rethinking data sharing and human participant protection in social science research: Applications from the qualitative realm"
While data sharing is becoming increasingly common in quantitative social inquiry, qualitative data are rarely shared. One factor inhibiting data sharing is a concern about human participant protections and privacy. Protecting the confidentiality and safety of research participants is a concern for both quantitative and qualitative researchers, but it raises specific concerns within the epistemic context of qualitative research. Thus, the applicability of emerging protection models from the quantitative realm must be carefully evaluated for application to the qualitative realm. At the same time, qualitative scholars already employ a variety of strategies for human-participant protection implicitly or informally during the research process. In this practice paper, we assess available strategies for protecting human participants and how they can be deployed. We describe a spectrum of possible data management options, such as de-identification and applying access controls, including some already employed by the Qualitative Data Repository (QDR) in tandem with its pilot depositors. (Full article...)
|
Featured article of the week: February 12–18:
"Handling metadata in a neurophysiology laboratory"
To date, non-reproducibility of neurophysiological research is a matter of intense discussion in the scientific community. A crucial component to enhance reproducibility is to comprehensively collect and store metadata, that is, all information about the experiment, the data, and the applied preprocessing steps on the data, such that they can be accessed and shared in a consistent and simple manner. However, the complexity of experiments, the highly specialized analysis workflows, and a lack of knowledge on how to make use of supporting software tools often overburden researchers to perform such a detailed documentation. For this reason, the collected metadata are often incomplete, incomprehensible for outsiders, or ambiguous. Based on our research experience in dealing with diverse datasets, we here provide conceptual and technical guidance to overcome the challenges associated with the collection, organization, and storage of metadata in a neurophysiology laboratory. (Full article...)
|
Featured article of the week: February 5–11:
"ISO 15189 accreditation: Navigation between quality management and patient safety"
Accreditation is a valuable resource for clinical laboratories, and the development of an international standard for their accreditation represented a milestone on the path towards improved quality and safety in laboratory medicine. The recent revision of the international standard, ISO 15189, has further strengthened its value not only for improving the quality system of a clinical laboratory but also for better answering the request for competence, focus on customers’ needs and ultimate value of laboratory services. Although in some countries more general standards such as ISO 9001 for quality systems or ISO 17025 for testing laboratories are still used, there is increasing recognition of the value of ISO 15189 as the most appropriate and useful standard for the accreditation of medical laboratories. In fact, only this international standard recognizes the importance of all steps of the total testing process, namely extra-analytical phases, the need to focus on technical competence in addition to quality systems, and the focus on customers’ needs. (Full article...)
|
Featured article of the week: January 29–February 4:
"Compliance culture or culture change? The role of funders in improving data management and sharing practice amongst researchers"
There is a wide and growing interest in promoting research data management (RDM) and research data sharing (RDS) from many stakeholders in the research enterprise. Funders are under pressure from activists, from government, and from the wider public agenda towards greater transparency and access to encourage, require, and deliver improved data practices from the researchers they fund.
Funders are responding to this, and to their own interest in improved practice, by developing and implementing policies on RDM and RDS. In this review we examine the state of funder policies, the process of implementation and available guidance to identify the challenges and opportunities for funders in developing policy and delivering on the aspirations for improved community practice, greater transparency and engagement, and enhanced impact. (Full article...)
|
Featured article of the week: January 22–28:
"A review of the role of public health informatics in healthcare"
Recognized as information intensive, healthcare requires timely, accurate information from many different sources generated by health information systems (HIS). With the availability of information technology in today's world and its integration in healthcare systems, the term “public health informatics (PHI)” was coined and used. The main focus of PHI is the use of information science and technology for promoting population health rather than individual health. PHI has a disease prevention rather than treatment focus in order to prevent a chain of events that leads to a disease's spread. Moreover, PHI often operates at the government level rather than in the private sector. This review article provides an overview of the field of PHI and compares paper-based surveillance system and public health information networks (PHIN). The current trends and future challenges of applying PHI systems in the Kingdom of Saudi Arabia (KSA) were also reported. (Full article...)
|
Featured article of the week: January 15–21:
"Preferred names, preferred pronouns, and gender identity in the electronic medical record and laboratory information system: Is pathology ready?"
Electronic medical records (EMRs) and laboratory information systems (LISs) commonly utilize patient identifiers such as legal name, sex, medical record number, and date of birth. There have been recommendations from some EMR working groups (e.g., the World Professional Association for Transgender Health) to include preferred name, pronoun preference, assigned sex at birth, and gender identity in the EMR. These practices are currently uncommon in the United States. There has been little published on the potential impact of these changes on pathology and LISs. We review the available literature and guidelines on the use of preferred name and gender identity on pathology, including data on changes in laboratory testing following gender transition treatments. We also describe pathology and clinical laboratory challenges in the implementation of preferred name at our institution. (Full article...)
|
Featured article of the week: January 8–14:
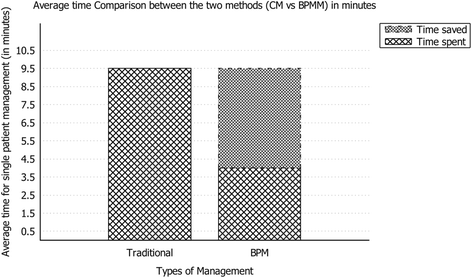
"Experimental application of business process management technology to manage clinical pathways: A pediatric kidney transplantation follow-up case"
Using a business process management platform, we implemented a specific application to manage the clinical pathway of pediatric patients, and we monitored the activities of the coordinator in charge of case management during a six-month period (from June 2015 to November 2015) using two methodologies: the traditional procedure and the one under study. The application helped physicians and nurses to optimize the amount of time and resources devoted to management purposes. In particular, time reduction was close to 60%. In addition, the reduction of data duplication, the integration of event management, and the efficient collection of data improved the quality of the service. The use of business process management technology, usually related to well-defined processes with high management costs, is an established procedure in multiple environments; its use in healthcare, however, is innovative. (Full article...)
|
Featured article of the week: January 1–7:
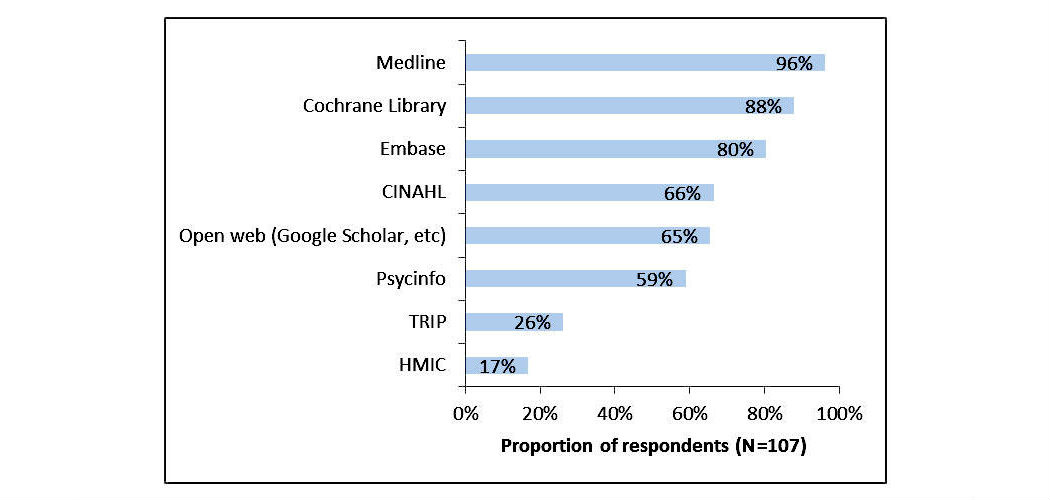
"Expert search strategies: The information retrieval practices of healthcare information professionals"
Healthcare information professionals play a key role in closing the knowledge gap between medical research and clinical practice. Their work involves meticulous searching of literature databases using complex search strategies that can consist of hundreds of keywords, operators, and ontology terms. This process is prone to error and can lead to inefficiency and bias if performed incorrectly.
The aim of this study was to investigate the search behavior of healthcare information professionals, uncovering their needs, goals, and requirements for information retrieval systems. A survey was distributed to healthcare information professionals via professional association email discussion lists. It investigated the search tasks they undertake, their techniques for search strategy formulation, their approaches to evaluating search results, and their preferred functionality for searching library-style databases. The popular literature search system PubMed was then evaluated to determine the extent to which their needs were met. (Full article...)
|
|