Journal:Application of informatics in cancer research and clinical practice: Opportunities and challenges
| Full article title | Application of informatics in cancer research and clinical practice: Opportunities and challenges |
|---|---|
| Journal | Cancer Innovation |
| Author(s) | Hong, Na; Sun, Gang; Zuo, Xiuran; Chen, Meng; Liu, Li; Jiani, Wang; Feng, Xiaobin; Shi, Wenzhao; Gong, Mengchun; Ma, Pengcheng |
| Author affiliation(s) | Digital Health China Technologies Co., Xinjiang Cancer Center, Huazhong University of Science and Technology, Southern Medical University, Chinese Academy of Medical Sciences and Peking Union Medical College, Tsinghua University |
| Primary contact | Email: gmc at nrdrs dot org |
| Year published | 2022 |
| Volume and issue | 1(1) |
| Page(s) | 80–91 |
| DOI | 10.1002/cai2.9 |
| ISSN | 2770-9183 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://onlinelibrary.wiley.com/doi/10.1002/cai2.9 |
| Download | https://onlinelibrary.wiley.com/doi/pdfdirect/10.1002/cai2.9 (PDF) |
Abstract
Cancer informatics has significantly progressed in the big data era. We summarize the application of informatics approaches to the cancer domain from both the informatics perspective (e.g., data management and data science) and the clinical perspective (e.g., cancer screening, risk assessment, diagnosis, treatment, and prognosis). We discuss various informatics methods and tools that are widely applied in cancer research and practices, such as cancer databases, data standards, terminologies, high-throughput omics data mining, machine learning algorithms, artificial intelligence imaging, and intelligent radiation. We also address the informatics challenges within the cancer field that pursue better treatment decisions and patient outcomes, and focus on how informatics can provide opportunities for cancer research and practices. Finally, we conclude that the interdisciplinary nature of cancer informatics and collaborations are major drivers for future research and applications in clinical practices. It is hoped that this review is instrumental for cancer researchers and clinicians with its informatics-specific insights.
Keywords: artificial intelligence application, cancer informatics, machine learning
Introduction
Advances in information science and technology have brought significant benefits to cancer research and care, including larger study cohorts, more complete follow-up, more effective clinician teams, lower costs, increased patient life expectancy, and improved quality of life. Despite all cancer-related aspects, such as diagnosis, prognosis, and treatment being significantly improved, this disease area remains one of the most significant challenges in medical science due to disease heterogeneity and the need to identify underlying biomarkers that are potentially linked to specific cancer types.
Cancer informatics is a branch of medical informatics that applies information science, computer science, data science, and information technologies to the field of oncology. This is an area that deals with the resources, devices, and methods required to optimize the acquisition, storage, retrieval, and use of information in cancer. Applied cancer informatics transforms clinical data into meaningful and useful information to improve processes and outcomes in patient-focused and evidence-based cancer care.[1] The fundamental goals of cancer informatics are: (1) to organize data in a way that is comprehensible and meaningful to clinicians, researchers, and patients; (2) to use data to advance cancer care and treatment; and (3) to yield new insights through data analysis.[2]
The multidisciplinary field of cancer informatics includes oncology, pathology, radiology, computational biology, physical chemistry, computer science, information systems, information management, biostatistics, clinical informatics, bioinformatics, imaging informatics, machine learning (ML), artificial intelligence (AI), data mining, data compliance, and many other disciplines. The integration and intersection of these individual disciplines bridge the gap between these individual cancer-related fields and promote cancer research and clinical practice.
From the point of view of informatics, methods and tools enhance the classification, accessibility, and applications of oncology data, thereby transforming cancer treatment into better outcomes. For example, with the development of clinical and imaging oncology databases, radiomics and AI have flourished, providing clinicians with a technological foundation for the early detection and treatment of cancer. In clinical practice, radiologists are under tremendous pressure as the number of cancer patients increases quickly. Studies in AI radiotherapy aim to make radiotherapy easier and faster and turn this labor-intensive procedure into a technology-intensive task. Another example is the multi-omics analysis of precision oncology. Multi-omics analyses can effectively overcome the limitations of single omics by integrating the analysis of a large amount of biological data at the molecular level in different dimensions, such as the genome, epigenome, transcriptome, proteome, metabolome, and microbiome. Moreover, it provides multi-level analyses and interpretations of complex life phenomena with many influencing factors, such as processes and diseases. With the popularization of next-generation high-throughput technologies and the accumulation of large amounts of multi-omics data, integration and fusion analysis for precise diagnosis and treatment of cancer has become an emerging trend.
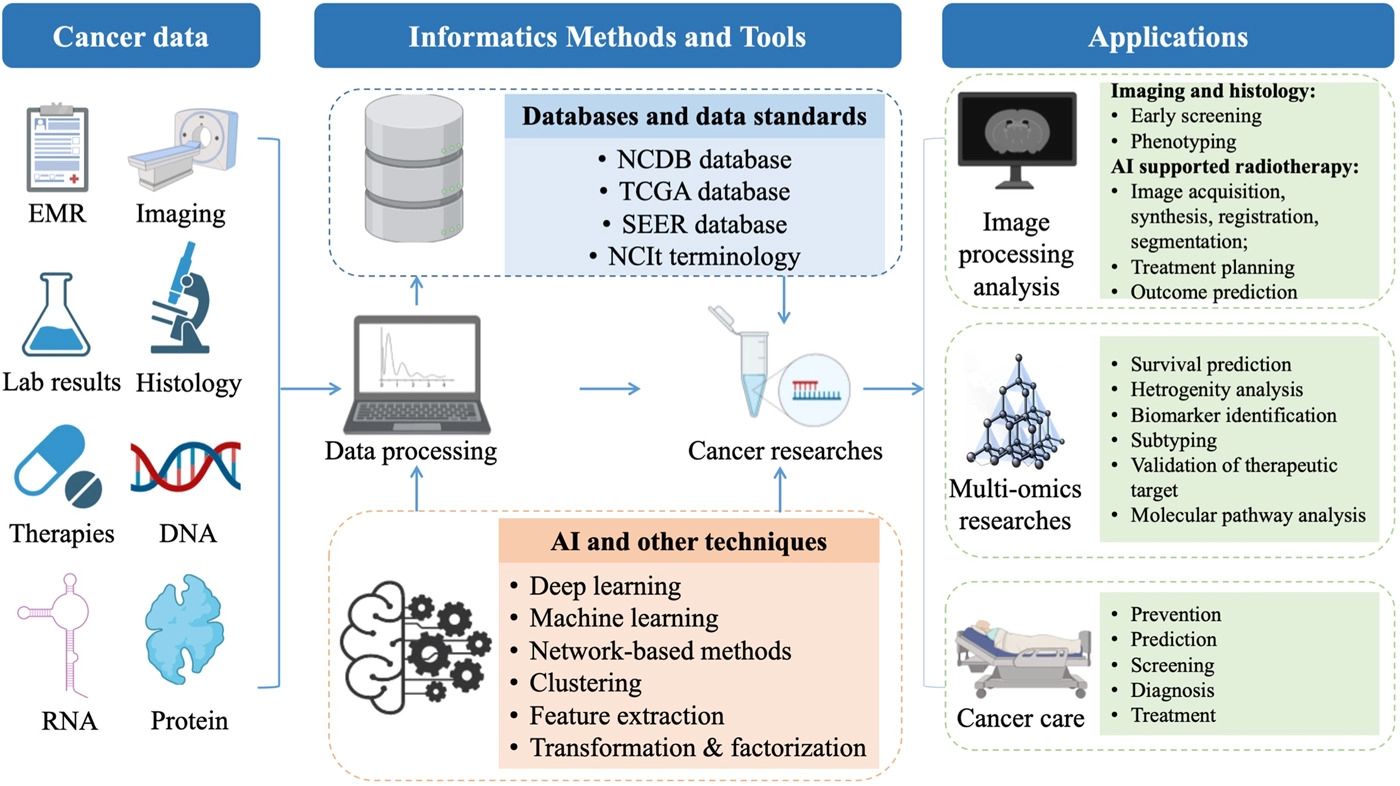
To summarize the current progress in informatics methods and tools to enhance cancer research and improve cancer clinical practices, we reviewed the most common recent scenarios of informatics-supported applications. A graphic abstract summarizing the field of cancer informatics is depicted in Figure 1.
|
Informatics-supported applications of cancer research and clinical practices
Informatics-based publications are available from the National Library of Medicine database (PubMed) and officially released web resources, which include cancer databases, cancer knowledge organization systems, cancer omics, and precision medicine, as well as AI-supported cancer imaging and radiotherapy. In this review, retrieved articles were manually screened according to a criterion containing the following items: aim of the study, methods, results, and clinical scenarios.
Databases and data standards for oncology
Healthcare data stored in various electronic systems follow different formats, whether structured or unstructured data. The information contained in medical records contains critical elements that support cancer therapies. Storing, extracting, and encoding such information plays an important role in cancer treatment and research. Population-based cancer registry databases can record information on incidence, mortality, and treatment outcomes, generating annual statistics as a result.[3] In contrast, hospital-based cancer databases provide more clinical information than population-based cancer registries, such as patient information, clinicopathological information, genomic data, disease staging, treatment, follow-up, lab test results, and medical records, which supports clinical research and improves the care of cancer patients.[3][4] Furthermore, a consistent system of coding needs to be ensured to integrate the collected data from different sources that could be encoded in various terminological standards.[5] In addition, ontology—as an integration of knowledge, annotation, and concepts—plays an important role in cancer treatment and research.
Cancer databases and scientific programs
The database built by the National Cancer Institute's (NCI's) Surveillance, Epidemiology, and End Results (SEER) program in 1973, and by the Centers for Disease Control and Prevention's (CDC's) National Program of Cancer Registries (NPCR) of the United States in 1995, is used to construct the US Cancer Statistics (USCS)[6][7], while data from the National Central Cancer Registry of China is used to produce cancer statistics in China.[8][9] The National Cancer Database (NCDB) of the United States is one of the largest cancer clinical registry databases, with over 34 million data sets of commonly diagnosed solid tumors added since 1989, and it has an increasing number of published studies.[10][11] Moreover, thousands of new genomes have been sequenced over the past few years.[12] The Cancer Genome Atlas (TCGA) was initiated in 2006 and has characterized more than 20,000 primary cancers at the molecular level, covering 33 cancer types to date. This database consists of genomic, expression, methylation, copy number variation, epigenomic, transcriptomic, and proteomic data amounting to more than 2.5 petabytes in volume.[13][14] The International Cancer Genome Consortium (ICGC) supports genomic studies in more than 50 cancer types involving more than 25,000 cancer genomes at the genomic, epigenomic, and transcriptomic levels.[15]
Cancer classification, terminology, and ontology
Cancer classification is the prime issue during patient treatment. The International Classification of Diseases for Oncology (ICD-O) published by the World Health Organization (WHO) is widely implemented for tumor disease classification. ICD-O uses a multi-axial coding system to classify the anatomical site and the histology of a tumor. The first, second, and third editions of ICD-O were published in 1976, 1991, and 2000, respectively.[16][17][18] Furthermore, the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) uses concepts, descriptions, and relationships to build terminology systems that can map and link to other standards.[19][20] It is used to encode cancer pathological checklists that aim to provide interoperable and portable diagnostic, prognostic, and predictive elements.[21][22] The NCI has published a comprehensive logic-based terminology, the National Cancer Institute Thesaurus (NCIt), covering cancer-related components such as clinical findings, drugs, treatments, anatomy, genes, proteins, and molecular information.[23] Adverse events (AEs), a critical element in cancer clinical trials and research, are recorded in dictionaries such as the Common Terminology Criteria for Adverse Events (CTCAE) and the Medical Dictionary for Regulatory Activities (MedDRA) developed by the NCI and the International Conference on Harmonization (ICH), respectively.[24][25] HemOnc.org—which contains information on drugs and regimens regarding their mechanism, U.S. Food and Drug Administration (FDA) approval, common usage, and synonyms—is published and maintained to meet the growing number of chemotherapeutic regimens by combining various definitions, such as RxNorm, SNOMED CT, and the NCIt.[19][26][27]
Cancer Care Treatment Outcome Ontology (CCTOO) describes treatment or trial endpoints for patients with solid tumors in four domains, 13 subgroups, and two concept hierarchical structures, with a total of 1,133 terms.[28] Alternatively, TNM-Ontology (TNM-O) consists of four parts: a representation of the primary tumor (T), a representation of regional lymph nodes (N), a representation of distant metastases (M), and the anatomical location of the tumor. It sets different T, N, and M code descriptors for tumors at different anatomical locations. TNM-O was implemented in a colorectal cancer database and achieved a 100% concordance rate after validation by experienced pathologists.[29] The Radiation Oncology Ontology (ROO) was published using Semantic Web technologies, forming a hierarchical structure containing 1,183 classes and 211 properties between classes[30], while the Radiation Oncology Structures (ROS) ontology was developed using a taxonomic hierarchy consisting of 417 classes, each with a number of subclasses, 81% of which can be mapped to the Unified Medical Language System (UMLS).[31] Cancer Cell Ontology (CCL) was published to represent cancer cell types via immune phenotypes in the field of hematological malignancies, with a total number of 6,900 classes (over 300 new classes added).[32] Prostate Cancer Ontology (PCO) represents integrated information from multiple prostate databases using a nine-level hierarchical structure, with 412 concepts[33] and local terminologies, such as the Cervical Cancer Common Terminology[34], which are used for supporting semantic interoperability and utilization of local clinical data.
AI-supported image processing and radiotherapy
Medical imaging is a useful and important modality for cancer detection, progression monitoring, and prognosis prediction. Radiomics and radiotherapy are the two most focused medical research and application areas advanced by AI. Radiomics refers to converting images into structured, mineable data.[35] Most AI-supported image applications focus on early screening and diagnosis using ML methods based on predefined features extracted from medical images.[36] Radiation therapy is a pivotal cancer treatment that has significantly progressed over the last decade due to numerous technological breakthroughs. Traditional radiation therapy workflows identify areas that would benefit from AI, including imaging, treatment planning, quality assurance, and outcome prediction. Many recent studies have shown that the adoption of radiomics and ML has paved the way for improved management of radiation therapy patients.
AI imaging and diagnostics
AI has contributed to medical imaging by improving the quality of images and computer-aided image interpretation and radiomics in most oncology-related diagnoses, and the application of AI is crucial in radiology for various modalities with improved quality, such as X-rays, ultrasounds, computed tomography, magnetic resonance imaging (MRI), positron emission tomography (PET), and digital pathology. To analyze these quantitative data, data images, predictive models, diagnosis, prognosis, and longitudinal monitoring based on a parsimonious set of informative imaging features are yielded. Images are analyzed with highly specialized algorithms with increased speed and accuracy.
According to a number of papers published in recent years, the most common cancer locations are the breast, kidney, brain, lung, prostate, cervix, and liver. The main AI algorithms are convolutional neural networks (CNNs), neural networks (NNs), support vector machines (SVMs), deep neural networks (DNNs), and ensemble learning techniques.[37] A recent study outlined the development and validation of an automated detection system for chest radiography with algorithms based on deep learning.[38] This automated system is designed to diagnose common thoracic diseases, including lung malignancies. The results of this study showed that AI-integrated systems have superior image recognition and analysis capabilities compared with human observers. For example, mammography is the first line of imaging screening for breast cancer. For younger women with dense breast tissue, ultrasound is the preferred option, and a previous study demonstrated the influence of AI in breast imaging.[39] The authors compared the interpretation of mammography with and without the assistance of AI. Unsurprisingly, radiologists with AI assistance were able to analyze mammography images quicker and more accurately, which is vital for the rapid detection of cancers, and further research directions for AI in medical imaging will focus on improving speed and reducing costs.[40][41] Previous studies have also reported AI tools developed by Google that can search for morphologically similar features[41], regardless of annotation status. For example, LYmph Node Assistant (LYNA) is a Google-developed deep learning algorithm that can successfully detect metastatic breast cancer on slides with up to 99% accuracy.
AI-supported radiotherapy
In radiotherapy, images from different patients, times, or modalities often need to be registered to synthesize their corresponding information in a joint coordinate. The registration of images is relatively simple. However, how to achieve the registration of images and pathology (biomarkers) obtained or analyzed by different modalities is a current problem. At present, the prediction of biomarkers according to images does not achieve accurate point-to-point matching. A study was conducted to set up the conditional Generative Adversarial Network (cGAN), which uses synthetic computed tomography (sCT) images from low field MR images in the pelvis and abdomen, and compares the differences in dose-volume histograms between sCT and original CT.[42] Deep learning has been used to improve the quality and efficiency of deformable image registration (DIR).[43] Given the unavoidable nonrigid anatomical motion by the patient between image acquisitions, DIR needs to establish a voxel-to-voxel correspondence between two medical images that reflects these two different anatomical instances.[44][45] In addition, treatment planning benefits from AI and information technologies. An array of research with dose prediction or validation has been published in recent years. Multiple dose levels, radiation-sensitive critical structures near target organs, and tumors in the abdomen, head, and neck were the most researched areas among recent achievements.[46] To enable accurate MRI-based dose calculations, Matteo et al. generated sCT from T1-weighted MRI using three 2D conditional cGANs.[47] Furthermore, new devices, such as electronic portal imaging devices (EPID)[48] and kV cone-beam computed tomography (CBCT) images[49], have reconstructed the 3D dose distribution in radiotherapy treatment. AI also supports radiotherapy outcome prediction, a dual-input channel hybrid deep learning model that efficiently integrates an entire set of dosimetric parameters for radiation treatment planning, which was developed to enhance the prediction of Grade 4 radiotherapy-induced lymphopenia.[50]
Cancer multi-omics research
Unlike evidence-based medicine, studies on precision oncology should be data-driven, and omics data are among the most critical. Omics is a type of biotechnology that analyzes the structure and function of the overall composition of a given biological function at different levels. With the development of high-throughput technologies, such as next-generation sequencing (NGS) and mass spectrometry (MS)-based techniques such as liquid chromatography–tandem mass spectrometry (LC-MS/MS), it is possible to facilitate the investigation of the genome, transcriptome, proteome, and metabolome. Compared with single-level omics, multi-omic approaches can reveal the molecular mechanisms underlying different phenotypic manifestations of cancer from multiple dimensions. Thus, multi-omics has been proposed as the key to precision oncology in clinical practice. Together, these omics data can help to reveal the complex molecular mechanisms in different diseases.[51] Multi-omics can generate more information, and how to achieve multi-omics registration deserves further research.
Genomics, proteomics, metabolomics, and microbiomics in cancer research
Scientists have identified several mutated cancer genes through DNA sequencing techniques, such as PIK3CA, EGFR, and HER2.[52][53][54] In recent years, the application of NGS for DNA sequencing, coupled with analytical methods, has enabled unprecedented speed and precision in decoding human genomes.[55] In addition, NGS techniques have dramatically reduced the cost of sequencing. Massively parallel sequencing allows further insights into cancer disease from various aspects, including diagnosis, classification, therapeutics, and risk prediction.[56] In addition to differences in gene expression, a study has suggested that DNA methylation, a reversible DNA modification, can be used as an indicator of cancer status.[57] The identification of DNA modifications—including methylation, acetylation, histone modification, and nucleosome remodeling—is defined as epigenomics. These modifications are critical in regulating the biological processes fundamental to cancer genesis.[58] Several factors such as genetic and environmental factors can affect DNA modifications, which might be long-lasting or even heritable.[59][60][61] Hence, epigenomics data has great potential in the interpretation of genetic variants in cancer. Compared with DNA, RNA molecules change temporally according to cellular, environmental, extracellular, and developmental stimulation. The application of NGS has also facilitated transcriptomics studies because we can identify both the presence and abundance of RNA transcripts in a genome-wide manner via RNA-sequencing.[62] Studies on transcriptomics have revealed characteristic gene expression signatures in various cancer types that can help in clinical decisions, including diagnosis, treatment choices, and disease management. Furthermore, several clinical trial findings have been applied to predict the prognosis of different cancers, such as breast and lung cancer.[63][64] Gene expression sequencing has also been extended to single cells, which enriches the data of cancer cells and helps us to understand cancer heterogeneity.[65][66]
In cancer research, proteomics data has contributed to the development of biomarkers in cancer identification, as well as classification, prediction of drug sensitivity, and identification of proteins that may mediate drug resistance in different cancer types.[67][68][69] The development of LC-MS/MS techniques has provided a platform for proteomic analysis, for example, supporting proteomic alterations in various cancer tissues. The application of LC-MS/MS can be extended to small molecules, which allows us to study metabolomics data. Compared with the omics mentioned above, metabolomics is a new field, and most studies of cancer metabolomics have focused on the identification of biomarkers in plasma or serum samples, such as unsaturated free fatty acids in colorectal cancer and citrate changes in prostate cancer.[70][71] Furthermore, microbiomics data give us brand new insights into cancer research and provide further information on the underlying molecular mechanisms in cancer genesis and development. It is suggested that the dysbiosis of symbiotic microbiota is related to several types of cancer.[72] In addition to cancer triggering or promotion, the microbiome can also be used in cancer therapies, including therapeutic targets and microbiota transplantation.[72][73]
Integrated multi-omics analysis for precision oncology
The integration and analysis of high-throughput omics data are complex but critical. Data-driven methods include deep learning, network-based methods, clustering, features extraction, transformation, and factorization, which connect the data and clinical and molecular features of cancer.[74] Furthermore, multi-omics studies on cancer cover many goals, including biomarker discovery, subgroup identification, molecular pathway analysis, and drug repurposing/discovery. Table 1 summarizes some multi-omics studies conducted on cancer in recent years. These findings have contributed to precision oncology in clinical decision-making and mechanism studies.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Biomarker identification for cancer prevention, diagnosis, and prognosis
Molecular biomarkers identified from omics data are often used for cancer prevention and diagnostics by detecting early disease. Cancer surveillance can be improved by identifying clinically relevant biomarkers for the early prevention of disease and to predict prognosis for effective treatment, such as carcinoembryonic antigen to monitor the recurrence of colorectal cancer[82][83] and mutations in estrogen receptor 1 (ESR1) to predict prognosis and treatment outcomes in breast cancer.[84] Furthermore, shallow sequencing has recently been applied to the whole genome for diagnostics in breast cancer[85], lung cancer[86], and neuroblastoma.[87]
Challenges
Driven by electronic and smart technologies, patient data are being generated at an increasingly rapid rate. However, regardless of the benefits of informatics, there are still many barriers to implementing AI in healthcare.
Data heterogeneity
The heterogeneity of cancer data is the primary difficulty in effectively integrating, searching, and extracting information, while the realization of its interoperability is a prerequisite for the implementation of personalized and precise treatment. Therefore, the inability to exchange information between cancer diagnosis and treatment systems becomes a limitation in pursuing data-driven clinical practice. Developing a global system to formalize and harmonize each individual data model, classification, thesaurus, vocabulary, terminology, and ontology from different systems is the main challenge.
Lack of good governance and annotated data
The limitations of most existing applications are the lack of quality control, data standardization, and sufficient samples. Most radiomics studies use images obtained from a wide range of scanning devices (e.g., CT, MRI, and PET) produced by different manufacturers. The absence of standardized protocols leads to significant variability in data acquisition and reconstruction parameters. Hence, numerous technical problems must be considered, and approved methodologies are needed to distinguish signal from noise in medical images[36], which requires the standardization of image preprocessing, tissue segmentation, feature calculation, and statistical methodologies.
Varying maturity of different informatics approaches in clinical application
The major challenge of informatics applications in cancer is the varying maturity of the different approaches. Genomics has been used for diagnosis, while other omics approaches such as epigenomics and proteomics are less used in clinical practice.[88] The time it takes to run the samples and the equipment requirements for omics data analysis techniques are variable. The technical maturity ranks from high to low as follows: RNA, epigenomics, transcriptomics, metabolomics, and proteomics. Furthermore, although data-driven analysis in cancer research is rapidly on the rise, most studies have focused on common cancer types, and there is still a lack of investigation of rare or challenging tumor types.[89] Imaging analysis technologies and tools are more mature than clinical data modeling and omics data analysis.
Model generalizability, results interpretation, and external validation
The generalizability of machine learning models is challenging. Different image acquisition equipment, different contrast agents, and different image acquisition parameters of the same equipment may have a large impact on the results. Furthermore, another challenge lies in the interpretation of data-driven results. Current AI predictions are more of a black box, and their interpretability and application are questioned by clinicians and require attention. This also brings challenges to the promotion of devices such as intelligent diagnosis. The data generated is only useful when it is clinically relevant and correctly interpreted. Thus, prospective clinical trials are urgently needed. All prospective studies with external validation are needed to translate these results from bench to bedside. However, both the scarcity of external data and the non-uniform method of external validation make this challenging.[90][91] Furthermore, to implement AI-based systems for routine clinical practice, the intended users require training and understanding of the system.[92]
Cost challenge
The implementation of informatics, AI, and data engineering approaches such as big data storage, curation, annotation, AI model training, and deployment requires enormous infrastructure, strong computing power, large storage capacity, massive multidisciplinary specifics, and time to integrate and interpret patient data. Informatics-supported application systems can be expensive because of their dependence on specialized computational requirements for fast data processing and rich medical knowledge for supporting medical applications appropriately. It is expected that advanced informatics methods and tools will reduce the cost, increase the speed of high-throughput data analysis, provide data services in a cost- and time-effective manner, and become widely accessible for cancer research and clinical applications.
Compliance challenge
Informatics systems process a huge amount of patient data, which could trigger the laws and regulations of data security for personal data protection, for example, the Personal Information Protection Law (PIPL) and Data Security Law (DSL) of China. How to protect patients' data and process sensitive data efficiently for the purposes of research as well as for clinical application is a challenge for medical institutions. Thus, a systematic approach for the purpose of compliance may apply to informatics practice.
Opportunities and future perspectives
Cancer burden is a global phenomenon. The reduction of mortality rates requires early diagnosis and effective therapeutic interventions. However, metastatic and recurrent cancers develop drug resistance. Thus, it is imperative to detect novel biomarkers that induce drug resistance and to identify therapeutic targets to improve treatment effects. Informatics methods and tools can be applied to several clinical applications, which are important for risk prediction, early detection of disease, diagnosis by sequencing and medical imaging, accurate prognosis, biomarker detection, and identification of therapeutic targets for novel drug discovery.
As a hierarchical structure with standardized concepts, data standards such as vocabularies, terminologies, and ontologies can promote tumor data integration in many aspects. As an easier and faster way to integrate and encode different data systems, vocabulary sharing and ontology matching can promote data communication between scientists and enable rapid information dissemination, thus facilitating the long-term evaluation of tumor treatment and research. A shared vocabulary standardizes the definition of data elements, which can make both humans and computers readable and accurately transmit information between systems and humans. Meanwhile, the semantic relationship between the data elements in an encoded system can also support the derivation of conclusions. Furthermore, ontology matching would entail establishing the relationships that exist between the terms of different ontologies. Therefore, it is beneficial to develop automatic mapping algorithms and ensure semantic consistency.
In addition, the application of high-throughput multi-omics data and mass spectrometry enable cancer researchers to perform large-scale studies to analyze the cellular/disease progression of various dimensions, from genome to proteome and metabolome. Furthermore, advanced methods and powerful computational tools will help to identify the links between the phenotypes and omics data. Multi-omics data platforms provide an opportunity to better understand cellular pathways in disease processes. Genomic analysis in cancer research has made significant progress in recent decades, and further studies will focus on RNA, protein, and metabolite changes and the role of the microbiome in disease. This systematic research on multi-level data can promote the development of prediction models and practical strategies for personalized cancer therapy.[62]
AI techniques, particularly ML, have been extensively applied to process large-scale and heterogeneous cancer data. These techniques have achieved good results in data mining and analysis by providing powerful algorithms. Therefore, future studies on cancer will be based on AI techniques to process not only structured clinical data but also other unstructured clinical data, such as electronic medical records (EMRs), imaging, and omics data. AI has made a significant impact and will continue to revolutionize healthcare and precision oncology. Considering the interdisciplinary nature of cancer informatics, the collaboration of multiple disciplines is a major driver for future research and applications.
Conclusion
In conclusion, clinical oncology and research are reaping the benefits of informatics. Using informatics methods and tools, a large amount of diverse and dynamic data plays an important role in cancer research and clinical practices in the workflow of data collection, modeling, interoperability, integration, analysis, and utilization. With the further development of convenient and intelligent tools, informatics will enable earlier cancer detection, more precise cancer treatment, and better outcomes.
Abbreviations, acronyms, and initialisms
- AE: adverse event
- AI: artificial intelligence
- CBCT: cone-beam computed tomography
- CCTOO: The Cancer Care Treatment Outcome Ontology
- CDC: Centers for Disease Control and Prevention
- cGAN: conditional Generative Adversarial Network
- CNN: convolutional neural network
- CT: computed tomography
- CTCAE: Common Terminology Criteria for Adverse Events
- DSL: Data Security Law
- DIR: deformable image registration
- DNN: deep neural network
- EMR: electronic medical record
- EPID: electronic portal imaging device
- ESR1: estrogen receptor 1
- ICD-O: International Classification of Diseases for Oncology
- ICGC: International Cancer Genome Consortium
- ICH: International Conference on Harmonization
- LYNA: LYmph Node Assistant
- MedDRA: Medical Dictionary for Regulatory Activities
- ML: machine learning
- MRI: magnetic resonance imaging
- MS: mass spectrometry
- NCCR: National Central Cancer Registry
- NCDB: National Cancer Database
- NCI: National Cancer Institute
- NCIt: National Cancer Institute Thesaurus
- NGS: next-generation sequencing
- NN: neural network
- NPCR: National Program of Cancer Registries
- PIPL: Personal Information Protection Law
- PCO: Prostate Cancer Ontology
- PET: positron-emission tomography
- ROO: Radiation Oncology Ontology
- ROS: Radiation Oncology Structures
- sCT: synthetic computed tomography
- SEER: Surveillance, Epidemiology, and End Results
- SNOMED CT: Systematized Nomenclature of Medicine Clinical Terms
- SVM: support vector machine
- TCGA: The Cancer Genome Atlas
- TNM-O: TNM-Ontology
- UMLS: Unified Medical Language System
- USCS: US Cancer Statistics
- WHO: World Health Organization
Acknowledgements
We thank Chao Liu, Ge Wu, Xiaoyu Wu, Yuanshi Jiao, and Yunchuan Qiao for their literature collection and review support.
Author contributions
Na Hong: conceptualization (equal); formal analysis (equal); investigation (equal); writing – original draft (equal); writing – review and editing (equal). Gang Sun: conceptualization (equal); formal analysis (equal); methodology (equal); writing – original draft (equal). Xiuran Zuo: investigation (equal); methodology (equal); resources (equal); writing – original draft (equal). Meng Chen: methodology (equal); writing – review and editing (equal). Li Liu: methodology (equal); writing – review and editing (equal). Jiani Wang: investigation (equal); writing – review and editing (equal). Xiaobin Feng: methodology (equal); writing – review and editing (equal). Wenzhao Shi: funding acquisition (equal); supervision (equal). Mengchun Gong: conceptualization (equal); funding acquisition (equal); project administration (equal). Pengcheng Ma: conceptualization (equal); funding acquisition (equal); writing – original draft (equal).
Conflict of interest
Professor Meng Chen and Mengchun Gong are members of Cancer Innovation Editorial Board. To minimize bias, they were excluded from all editorial decision-making related to the acceptance of this article for publication. The remaining authors declare no conflict of interest.
References
- ↑ National Cancer Registrars Association. "Cancer Informatics". https://www.ncra-usa.org/Education/Informatics.
- ↑ Warner, Jeremy L.; Patt, Debra; Section Editors for the IMIA Yearbook Section on Cancer Informatics (1 August 2020). "Cancer Informatics in 2019: Deep Learning Takes Center Stage" (in en). Yearbook of Medical Informatics 29 (01): 243–246. doi:10.1055/s-0040-1701993. ISSN 0943-4747. PMC PMC7442504. PMID 32823323. http://www.thieme-connect.de/DOI/DOI?10.1055/s-0040-1701993.
- ↑ 3.0 3.1 Boffa, Daniel J.; Rosen, Joshua E.; Mallin, Katherine; Loomis, Ashley; Gay, Greer; Palis, Bryan; Thoburn, Kathleen; Gress, Donna et al. (1 December 2017). "Using the National Cancer Database for Outcomes Research: A Review" (in en). JAMA Oncology 3 (12): 1722–8. doi:10.1001/jamaoncol.2016.6905. ISSN 2374-2437. http://oncology.jamanetwork.com/article.aspx?doi=10.1001/jamaoncol.2016.6905.
- ↑ McCabe, Ryan M. (1 October 2019). "National Cancer Database: The Past, Present, and Future of the Cancer Registry and Its Efforts to Improve the Quality of Cancer Care" (in en). Seminars in Radiation Oncology 29 (4): 323–325. doi:10.1016/j.semradonc.2019.05.005. https://linkinghub.elsevier.com/retrieve/pii/S1053429619300372.
- ↑ Jouhet, V.; Defossez, G.; Ingrand, P.; CRISAP; CoRIM (2013). "Automated Selection of Relevant Information for Notification of Incident Cancer Cases within a Multisource Cancer Registry" (in en). Methods of Information in Medicine 52 (05): 411–421. doi:10.3414/ME12-01-0101. ISSN 0026-1270. http://www.thieme-connect.de/DOI/DOI?10.3414/ME12-01-0101.
- ↑ Siegel, Rebecca L.; Miller, Kimberly D.; Fuchs, Hannah E.; Jemal, Ahmedin (1 January 2022). "Cancer statistics, 2022" (in en). CA: A Cancer Journal for Clinicians 72 (1): 7–33. doi:10.3322/caac.21708. ISSN 0007-9235. https://onlinelibrary.wiley.com/doi/10.3322/caac.21708.
- ↑ Islami, Farhad; Ward, Elizabeth M; Sung, Hyuna; Cronin, Kathleen A; Tangka, Florence K L; Sherman, Recinda L; Zhao, Jingxuan; Anderson, Robert N et al. (29 November 2021). "Annual Report to the Nation on the Status of Cancer, Part 1: National Cancer Statistics" (in en). JNCI: Journal of the National Cancer Institute 113 (12): 1648–1669. doi:10.1093/jnci/djab131. ISSN 0027-8874. PMC PMC8634503. PMID 34240195. https://academic.oup.com/jnci/article/113/12/1648/6312532.
- ↑ Chen, Wanqing; Zheng, Rongshou; Baade, Peter D.; Zhang, Siwei; Zeng, Hongmei; Bray, Freddie; Jemal, Ahmedin; Yu, Xue Qin et al. (1 March 2016). "Cancer statistics in China, 2015: Cancer Statistics in China, 2015" (in en). CA: A Cancer Journal for Clinicians 66 (2): 115–132. doi:10.3322/caac.21338. http://doi.wiley.com/10.3322/caac.21338.
- ↑ Wei, Wenqiang; Zeng, Hongmei; Zheng, Rongshou; Zhang, Siwei; An, Lan; Chen, Ru; Wang, Shaoming; Sun, Kexin et al. (1 July 2020). "Cancer registration in China and its role in cancer prevention and control" (in en). The Lancet Oncology 21 (7): e342–e349. doi:10.1016/S1470-2045(20)30073-5. https://linkinghub.elsevier.com/retrieve/pii/S1470204520300735.
- ↑ American College of Surgeons. "National Cancer Database". Quality Programs. https://www.facs.org/quality-programs/cancer-programs/national-cancer-database/.
- ↑ Blanchard, Pierre; Garden, Adam S. (1 July 2016). "Looking Beyond the Numbers: Highlighting the Challenges of Population-Based Studies in Cancer Research" (in en). Journal of Clinical Oncology 34 (19): 2317–2318. doi:10.1200/JCO.2015.66.0894. ISSN 0732-183X. https://ascopubs.org/doi/10.1200/JCO.2015.66.0894.
- ↑ Telenti, Amalio; Pierce, Levi C. T.; Biggs, William H.; di Iulio, Julia; Wong, Emily H. M.; Fabani, Martin M.; Kirkness, Ewen F.; Moustafa, Ahmed et al. (18 October 2016). "Deep sequencing of 10,000 human genomes" (in en). Proceedings of the National Academy of Sciences 113 (42): 11901–11906. doi:10.1073/pnas.1613365113. ISSN 0027-8424. PMC PMC5081584. PMID 27702888. https://pnas.org/doi/full/10.1073/pnas.1613365113.
- ↑ Wang, Zhining; Jensen, Mark A.; Zenklusen, Jean Claude (2016), Mathé, Ewy; Davis, Sean, eds., "A Practical Guide to The Cancer Genome Atlas (TCGA)" (in en), Statistical Genomics (New York, NY: Springer New York) 1418: 111–141, doi:10.1007/978-1-4939-3578-9_6, ISBN 978-1-4939-3576-5, http://link.springer.com/10.1007/978-1-4939-3578-9_6. Retrieved 2022-12-06
- ↑ National Cancer Institute. "The Cancer Genome Atlas Program". U.S. Department of Health and Human Services. https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga.
- ↑ The International Cancer Genome Consortium (15 April 2010). "International network of cancer genome projects" (in en). Nature 464 (7291): 993–998. doi:10.1038/nature08987. ISSN 0028-0836. PMC PMC2902243. PMID 20393554. http://www.nature.com/articles/nature08987.
- ↑ Percy, Constance L.; Van Holten, Valerie; Muir, C. S., eds. (1990). International classification of diseases for oncology =: ICD-O (2nd ed ed.). Geneva: World Health Organization. ISBN 978-92-4-154414-6.
- ↑ Fritz, April G., ed. (2013). International classification of diseases for oncology: ICD-O (Third edition, First revision ed.). Geneva: World Health Organization. ISBN 978-92-4-154849-6.
- ↑ Fritz, April G., ed. (2000). International classification of diseases for oncology: ICD-O (3rd ed ed.). Geneva: World Health Organization. ISBN 978-92-4-154534-1. OCLC ocm45716980. https://www.worldcat.org/title/mediawiki/oclc/ocm45716980.
- ↑ 19.0 19.1 SNOMED International. "The Value of SNOMED CT". https://www.snomed.org/snomed-ct/why-snomed-ct.
- ↑ Nikiema, Jean Noël; Jouhet, Vianney; Mougin, Fleur (1 October 2017). "Integrating cancer diagnosis terminologies based on logical definitions of SNOMED CT concepts" (in en). Journal of Biomedical Informatics 74: 46–58. doi:10.1016/j.jbi.2017.08.013. https://linkinghub.elsevier.com/retrieve/pii/S153204641730196X.
- ↑ Van Berkum, Monique M. (2003). "SNOMED CT encoded Cancer Protocols". AMIA ... Annual Symposium proceedings. AMIA Symposium 2003: 1039. ISSN 1942-597X. PMC 1480015. PMID 14728542. https://pubmed.ncbi.nlm.nih.gov/14728542.
- ↑ Torous, Vanda F.; Allan, Robert W.; Balani, Jyoti; Baskovich, Brett; Birdsong, George G.; Dellers, Elizabeth; Dryden, Mignon; Edgerton, Mary E. et al. (1 April 2021). "Exploring the College of American Pathologists Electronic Cancer Checklists: What They Are and What They Can Do for You" (in en). Archives of Pathology & Laboratory Medicine 145 (4): 392–398. doi:10.5858/arpa.2020-0239-ED. ISSN 1543-2165. https://meridian.allenpress.com/aplm/article/145/4/392/448764/Exploring-the-College-of-American-Pathologists.
- ↑ Sioutos, Nicholas; Coronado, Sherri de; Haber, Margaret W.; Hartel, Frank W.; Shaiu, Wen-Ling; Wright, Lawrence W. (1 February 2007). "NCI Thesaurus: A semantic model integrating cancer-related clinical and molecular information" (in en). Journal of Biomedical Informatics 40 (1): 30–43. doi:10.1016/j.jbi.2006.02.013. https://linkinghub.elsevier.com/retrieve/pii/S1532046406000311.
- ↑ National Cancer Institute (2006). CTEP: NCI Guidance on CTC Terminology Applications. National Cancer Institute.
- ↑ "Introductory Guide for Standardised MedDRA Queries (SMQs) Version 21.0." (PDF). International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use. March 2018. https://admin.meddra.org/sites/default/files/guidance/file/smq_intguide_21_0_english.pdf.
- ↑ Warner, Jeremy L.; Cowan, Andrew J.; Hall, Aric C.; Yang, Peter C. (1 May 2015). "HemOnc.org: A Collaborative Online Knowledge Platform for Oncology Professionals" (in en). Journal of Oncology Practice 11 (3): e336–e350. doi:10.1200/JOP.2014.001511. ISSN 1554-7477. PMC PMC5706141. PMID 25736385. https://ascopubs.org/doi/10.1200/JOP.2014.001511.
- ↑ "RxNorm". Unified Medical Language System. National Library of Medicine. 2022. https://www.nlm.nih.gov/research/umls/rxnorm/index.html.
- ↑ Lin, Frank P.; Groza, Tudor; Kocbek, Simon; Antezana, Erick; Epstein, Richard J. (1 December 2018). "Cancer Care Treatment Outcome Ontology: A Novel Computable Ontology for Profiling Treatment Outcomes in Patients With Solid Tumors" (in en). JCO Clinical Cancer Informatics (2): 1–14. doi:10.1200/CCI.18.00026. ISSN 2473-4276. https://ascopubs.org/doi/10.1200/CCI.18.00026.
- ↑ Boeker, Martin; França, Fábio; Bronsert, Peter; Schulz, Stefan (1 December 2016). "TNM-O: ontology support for staging of malignant tumours" (in en). Journal of Biomedical Semantics 7 (1): 64. doi:10.1186/s13326-016-0106-9. ISSN 2041-1480. PMC PMC5109740. PMID 27842575. http://jbiomedsem.biomedcentral.com/articles/10.1186/s13326-016-0106-9.
- ↑ Traverso, Alberto; van Soest, Johan; Wee, Leonard; Dekker, Andre (1 October 2018). "The radiation oncology ontology (ROO): Publishing linked data in radiation oncology using semantic web and ontology techniques" (in en). Medical Physics 45 (10): e854–e862. doi:10.1002/mp.12879. https://onlinelibrary.wiley.com/doi/10.1002/mp.12879.
- ↑ Bibault, Jean-Emmanuel; Zapletal, Eric; Rance, Bastien; Giraud, Philippe; Burgun, Anita (19 January 2018). Amendola, Roberto. ed. "Labeling for Big Data in radiation oncology: The Radiation Oncology Structures ontology" (in en). PLOS ONE 13 (1): e0191263. doi:10.1371/journal.pone.0191263. ISSN 1932-6203. PMC PMC5774757. PMID 29351341. https://dx.plos.org/10.1371/journal.pone.0191263.
- ↑ Serra, Lucas M.; Duncan, William D.; Diehl, Alexander D. (1 April 2019). "An ontology for representing hematologic malignancies: the cancer cell ontology" (in en). BMC Bioinformatics 20 (S5): 181. doi:10.1186/s12859-019-2722-8. ISSN 1471-2105. PMC PMC6509834. PMID 31272372. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-019-2722-8.
- ↑ Min, Hua; Manion, Frank J.; Goralczyk, Elizabeth; Wong, Yu-Ning; Ross, Eric; Beck, J. Robert (1 December 2009). "Integration of prostate cancer clinical data using an ontology" (in en). Journal of Biomedical Informatics 42 (6): 1035–1045. doi:10.1016/j.jbi.2009.05.007. PMC PMC2784120. PMID 19497389. https://linkinghub.elsevier.com/retrieve/pii/S1532046409000793.
- ↑ Hong, Na; Chang, Fengxiang; Ou, Zhengjie; Wang, Yishang; Yang, Yating; Guo, Qiang; Ma, Jianhui; Zhao, Dan (1 November 2021). "Construction of the cervical cancer common terminology for promoting semantic interoperability and utilization of Chinese clinical data" (in en). BMC Medical Informatics and Decision Making 21 (S9): 309. doi:10.1186/s12911-021-01672-x. ISSN 1472-6947. PMC PMC8596900. PMID 34789237. https://bmcmedinformdecismak.biomedcentral.com/articles/10.1186/s12911-021-01672-x.
- ↑ Gillies, Robert J.; Kinahan, Paul E.; Hricak, Hedvig (1 February 2016). "Radiomics: Images Are More than Pictures, They Are Data" (in en). Radiology 278 (2): 563–577. doi:10.1148/radiol.2015151169. ISSN 0033-8419. PMC PMC4734157. PMID 26579733. http://pubs.rsna.org/doi/10.1148/radiol.2015151169.
- ↑ 36.0 36.1 Reuzé, Sylvain; Schernberg, Antoine; Orlhac, Fanny; Sun, Roger; Chargari, Cyrus; Dercle, Laurent; Deutsch, Eric; Buvat, Irène et al. (1 November 2018). "Radiomics in Nuclear Medicine Applied to Radiation Therapy: Methods, Pitfalls, and Challenges" (in en). International Journal of Radiation Oncology*Biology*Physics 102 (4): 1117–1142. doi:10.1016/j.ijrobp.2018.05.022. https://linkinghub.elsevier.com/retrieve/pii/S0360301618308150.
- ↑ Kumar, Yogesh; Gupta, Surbhi; Singla, Ruchi; Hu, Yu-Chen (1 June 2022). "A Systematic Review of Artificial Intelligence Techniques in Cancer Prediction and Diagnosis" (in en). Archives of Computational Methods in Engineering 29 (4): 2043–2070. doi:10.1007/s11831-021-09648-w. ISSN 1134-3060. PMC PMC8475374. PMID 34602811. https://link.springer.com/10.1007/s11831-021-09648-w.
- ↑ Hwang, Eui Jin; Park, Sunggyun; Jin, Kwang-Nam; Kim, Jung Im; Choi, So Young; Lee, Jong Hyuk; Goo, Jin Mo; Aum, Jaehong et al. (22 March 2019). "Development and Validation of a Deep Learning–Based Automated Detection Algorithm for Major Thoracic Diseases on Chest Radiographs" (in en). JAMA Network Open 2 (3): e191095. doi:10.1001/jamanetworkopen.2019.1095. ISSN 2574-3805. PMC PMC6583308. PMID 30901052. http://jamanetworkopen.jamanetwork.com/article.aspx?doi=10.1001/jamanetworkopen.2019.1095.
- ↑ Rodríguez-Ruiz, Alejandro; Krupinski, Elizabeth; Mordang, Jan-Jurre; Schilling, Kathy; Heywang-Köbrunner, Sylvia H.; Sechopoulos, Ioannis; Mann, Ritse M. (1 February 2019). "Detection of Breast Cancer with Mammography: Effect of an Artificial Intelligence Support System" (in en). Radiology 290 (2): 305–314. doi:10.1148/radiol.2018181371. ISSN 0033-8419. http://pubs.rsna.org/doi/10.1148/radiol.2018181371.
- ↑ Lewis, Sarah J; Gandomkar, Ziba; Brennan, Patrick C (1 December 2019). "Artificial Intelligence in medical imaging practice: looking to the future" (in en). Journal of Medical Radiation Sciences 66 (4): 292–295. doi:10.1002/jmrs.369. ISSN 2051-3895. PMC PMC6920680. PMID 31709775. https://onlinelibrary.wiley.com/doi/10.1002/jmrs.369.
- ↑ 41.0 41.1 Gore, John C. (1 May 2020). "Artificial intelligence in medical imaging" (in en). Magnetic Resonance Imaging 68: A1–A4. doi:10.1016/j.mri.2019.12.006. https://linkinghub.elsevier.com/retrieve/pii/S0730725X19307556.
- ↑ Cusumano, Davide; Lenkowicz, Jacopo; Votta, Claudio; Boldrini, Luca; Placidi, Lorenzo; Catucci, Francesco; Dinapoli, Nicola; Antonelli, Marco Valerio et al. (1 December 2020). "A deep learning approach to generate synthetic CT in low field MR-guided adaptive radiotherapy for abdominal and pelvic cases" (in en). Radiotherapy and Oncology 153: 205–212. doi:10.1016/j.radonc.2020.10.018. https://linkinghub.elsevier.com/retrieve/pii/S0167814020308549.
- ↑ Balakrishnan, Guha; Zhao, Amy; Sabuncu, Mert R.; Guttag, John; Dalca, Adrian V. (1 August 2019). "VoxelMorph: A Learning Framework for Deformable Medical Image Registration". IEEE Transactions on Medical Imaging 38 (8): 1788–1800. doi:10.1109/TMI.2019.2897538. ISSN 0278-0062. https://ieeexplore.ieee.org/document/8633930/.
- ↑ Han, Xiao; Hoogeman, Mischa S.; Levendag, Peter C.; Hibbard, Lyndon S.; Teguh, David N.; Voet, Peter; Cowen, Andrew C.; Wolf, Theresa K. (2008), Metaxas, Dimitris; Axel, Leon; Fichtinger, Gabor et al.., eds., "Atlas-Based Auto-segmentation of Head and Neck CT Images", Medical Image Computing and Computer-Assisted Intervention – MICCAI 2008 (Berlin, Heidelberg: Springer Berlin Heidelberg) 5242: 434–441, doi:10.1007/978-3-540-85990-1_52, ISBN 978-3-540-85989-5, http://link.springer.com/10.1007/978-3-540-85990-1_52. Retrieved 2022-12-06
- ↑ Bondiau, Pierre-Yves; Malandain, Grégoire; Chanalet, Stéphane; Marcy, Pierre-Yves; Habrand, Jean-Louis; Fauchon, François; Paquis, Philippe; Courdi, Adel et al. (1 January 2005). "Atlas-based automatic segmentation of MR images: Validation study on the brainstem in radiotherapy context" (in en). International Journal of Radiation Oncology*Biology*Physics 61 (1): 289–298. doi:10.1016/j.ijrobp.2004.08.055. https://linkinghub.elsevier.com/retrieve/pii/S0360301604026823.
- ↑ Florkow, Mateusz C.; Guerreiro, Filipa; Zijlstra, Frank; Seravalli, Enrica; Janssens, Geert O.; Maduro, John H.; Knopf, Antje C.; Castelein, René M. et al. (1 December 2020). "Deep learning-enabled MRI-only photon and proton therapy treatment planning for paediatric abdominal tumours" (in en). Radiotherapy and Oncology 153: 220–227. doi:10.1016/j.radonc.2020.09.056. https://linkinghub.elsevier.com/retrieve/pii/S0167814020308318.
- ↑ Maspero, Matteo; Bentvelzen, Laura G.; Savenije, Mark H.F.; Guerreiro, Filipa; Seravalli, Enrica; Janssens, Geert O.; van den Berg, Cornelis A.T.; Philippens, Marielle E.P. (1 December 2020). "Deep learning-based synthetic CT generation for paediatric brain MR-only photon and proton radiotherapy" (in en). Radiotherapy and Oncology 153: 197–204. doi:10.1016/j.radonc.2020.09.029. https://linkinghub.elsevier.com/retrieve/pii/S0167814020308045.
- ↑ Jia, Mengyu; Wu, Yan; Yang, Yong; Wang, Lei; Chuang, Cynthia; Han, Bin; Xing, Lei (1 November 2021). "Deep learning‐enabled EPID‐based 3D dosimetry for dose verification of step‐and‐shoot radiotherapy" (in en). Medical Physics 48 (11): 6810–6819. doi:10.1002/mp.15218. ISSN 0094-2405. https://onlinelibrary.wiley.com/doi/10.1002/mp.15218.
- ↑ Barateau, Anaïs; De Crevoisier, Renaud; Largent, Axel; Mylona, Eugenia; Perichon, Nicolas; Castelli, Joël; Chajon, Enrique; Acosta, Oscar et al. (1 October 2020). "Comparison of CBCT‐based dose calculation methods in head and neck cancer radiotherapy: from Hounsfield unit to density calibration curve to deep learning" (in en). Medical Physics 47 (10): 4683–4693. doi:10.1002/mp.14387. ISSN 0094-2405. https://onlinelibrary.wiley.com/doi/10.1002/mp.14387.
- ↑ Zhu, Cong; Lin, Steven H; Jiang, Xiaoqian; Xiang, Yang; Belal, Zayne; Jun, Goo; Mohan, Radhe (4 February 2020). "A novel deep learning model using dosimetric and clinical information for grade 4 radiotherapy-induced lymphopenia prediction". Physics in Medicine & Biology 65 (3): 035014. doi:10.1088/1361-6560/ab63b6. ISSN 1361-6560. PMC PMC7501732. PMID 31851954. https://iopscience.iop.org/article/10.1088/1361-6560/ab63b6.
- ↑ Hasin, Yehudit; Seldin, Marcus; Lusis, Aldons (1 December 2017). "Multi-omics approaches to disease" (in en). Genome Biology 18 (1): 83. doi:10.1186/s13059-017-1215-1. ISSN 1474-760X. PMC PMC5418815. PMID 28476144. https://genomebiology.biomedcentral.com/articles/10.1186/s13059-017-1215-1.
- ↑ Samuels, Yardena; Wang, Zhenghe; Bardelli, Alberto; Silliman, Natalie; Ptak, Janine; Szabo, Steve; Yan, Hai; Gazdar, Adi et al. (23 April 2004). "High Frequency of Mutations of the PIK3CA Gene in Human Cancers" (in en). Science 304 (5670): 554–554. doi:10.1126/science.1096502. ISSN 0036-8075. https://www.science.org/doi/10.1126/science.1096502.
- ↑ Paez, J. Guillermo; Jänne, Pasi A.; Lee, Jeffrey C.; Tracy, Sean; Greulich, Heidi; Gabriel, Stacey; Herman, Paula; Kaye, Frederic J. et al. (4 June 2004). "EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy" (in en). Science 304 (5676): 1497–1500. doi:10.1126/science.1099314. ISSN 0036-8075. https://www.science.org/doi/10.1126/science.1099314.
- ↑ Stephens, Philip; Hunter, Chris; Bignell, Graham; Edkins, Sarah; Davies, Helen; Teague, Jon; Stevens, Claire; O'Meara, Sarah et al. (1 September 2004). "Intragenic ERBB2 kinase mutations in tumours" (in en). Nature 431 (7008): 525–526. doi:10.1038/431525b. ISSN 0028-0836. http://www.nature.com/articles/431525b.
- ↑ Mardis, Elaine R (1 June 2012). "Genome sequencing and cancer" (in en). Current Opinion in Genetics & Development 22 (3): 245–250. doi:10.1016/j.gde.2012.03.005. PMC PMC3890425. PMID 22534183. https://linkinghub.elsevier.com/retrieve/pii/S0959437X12000433.
- ↑ Kamps, Rick; Brandão, Rita; Bosch, Bianca; Paulussen, Aimee; Xanthoulea, Sofia; Blok, Marinus; Romano, Andrea (31 January 2017). "Next-Generation Sequencing in Oncology: Genetic Diagnosis, Risk Prediction and Cancer Classification" (in en). International Journal of Molecular Sciences 18 (2): 308. doi:10.3390/ijms18020308. ISSN 1422-0067. PMC PMC5343844. PMID 28146134. http://www.mdpi.com/1422-0067/18/2/308.
- ↑ Baylin, S. B. (1 April 2001). "Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer". Human Molecular Genetics 10 (7): 687–692. doi:10.1093/hmg/10.7.687. https://academic.oup.com/hmg/article-lookup/doi/10.1093/hmg/10.7.687.
- ↑ Dawson, Mark A.; Kouzarides, Tony (1 July 2012). "Cancer Epigenetics: From Mechanism to Therapy" (in en). Cell 150 (1): 12–27. doi:10.1016/j.cell.2012.06.013. https://linkinghub.elsevier.com/retrieve/pii/S0092867412007623.
- ↑ Gut, Philipp; Verdin, Eric (1 October 2013). "The nexus of chromatin regulation and intermediary metabolism" (in en). Nature 502 (7472): 489–498. doi:10.1038/nature12752. ISSN 0028-0836. http://www.nature.com/articles/nature12752.
- ↑ Liu, Liang; Li, Yuanyuan; Tollefsbol, Trygve O. (2008). "Gene-environment interactions and epigenetic basis of human diseases". Current Issues in Molecular Biology 10 (1-2): 25–36. ISSN 1467-3037. PMC 2434999. PMID 18525104. https://pubmed.ncbi.nlm.nih.gov/18525104.
- ↑ Taudt, Aaron; Colomé-Tatché, Maria; Johannes, Frank (1 June 2016). "Genetic sources of population epigenomic variation" (in en). Nature Reviews Genetics 17 (6): 319–332. doi:10.1038/nrg.2016.45. ISSN 1471-0056. http://www.nature.com/articles/nrg.2016.45.
- ↑ 62.0 62.1 Chakraborty, Sajib; Hosen, Md. Ismail; Ahmed, Musaddeque; Shekhar, Hossain Uddin (3 October 2018). "Onco-Multi-OMICS Approach: A New Frontier in Cancer Research" (in en). BioMed Research International 2018: 1–14. doi:10.1155/2018/9836256. ISSN 2314-6133. PMC PMC6192166. PMID 30402498. https://www.hindawi.com/journals/bmri/2018/9836256/.
- ↑ Prat, Aleix; Ellis, Matthew J.; Perou, Charles M. (1 January 2012). "Practical implications of gene-expression-based assays for breast oncologists" (in en). Nature Reviews Clinical Oncology 9 (1): 48–57. doi:10.1038/nrclinonc.2011.178. ISSN 1759-4774. PMC PMC3703639. PMID 22143140. http://www.nature.com/articles/nrclinonc.2011.178.
- ↑ Botling, Johan; Edlund, Karolina; Lohr, Miriam; Hellwig, Birte; Holmberg, Lars; Lambe, Mats; Berglund, Anders; Ekman, Simon et al. (1 January 2013). "Biomarker Discovery in Non–Small Cell Lung Cancer: Integrating Gene Expression Profiling, Meta-analysis, and Tissue Microarray Validation" (in en). Clinical Cancer Research 19 (1): 194–204. doi:10.1158/1078-0432.CCR-12-1139. ISSN 1078-0432. https://aacrjournals.org/clincancerres/article/19/1/194/77776/Biomarker-Discovery-in-Non-Small-Cell-Lung-Cancer.
- ↑ Song, Qianqian; Hawkins, Gregory A.; Wudel, Leonard; Chou, Ping‐Chieh; Forbes, Elizabeth; Pullikuth, Ashok K.; Liu, Liang; Jin, Guangxu et al. (1 June 2019). "Dissecting intratumoral myeloid cell plasticity by single cell RNA‐seq" (in en). Cancer Medicine 8 (6): 3072–3085. doi:10.1002/cam4.2113. ISSN 2045-7634. PMC PMC6558497. PMID 31033233. https://onlinelibrary.wiley.com/doi/10.1002/cam4.2113.
- ↑ Suvà, Mario L.; Tirosh, Itay (1 July 2019). "Single-Cell RNA Sequencing in Cancer: Lessons Learned and Emerging Challenges" (in en). Molecular Cell 75 (1): 7–12. doi:10.1016/j.molcel.2019.05.003. https://linkinghub.elsevier.com/retrieve/pii/S1097276519303569.
- ↑ Swiatly, Agata; Horala, Agnieszka; Matysiak, Jan; Hajduk, Joanna; Nowak-Markwitz, Ewa; Kokot, Zenon (31 July 2018). "Understanding Ovarian Cancer: iTRAQ-Based Proteomics for Biomarker Discovery" (in en). International Journal of Molecular Sciences 19 (8): 2240. doi:10.3390/ijms19082240. ISSN 1422-0067. PMC PMC6121953. PMID 30065196. http://www.mdpi.com/1422-0067/19/8/2240.
- ↑ Yanovich, Gali; Agmon, Hadar; Harel, Michal; Sonnenblick, Amir; Peretz, Tamar; Geiger, Tamar (15 October 2018). "Clinical Proteomics of Breast Cancer Reveals a Novel Layer of Breast Cancer Classification" (in en). Cancer Research 78 (20): 6001–6010. doi:10.1158/0008-5472.CAN-18-1079. ISSN 0008-5472. PMC PMC6193543. PMID 30154156. https://aacrjournals.org/cancerres/article/78/20/6001/631739/Clinical-Proteomics-of-Breast-Cancer-Reveals-a.
- ↑ Ali, Mehreen; Khan, Suleiman A; Wennerberg, Krister; Aittokallio, Tero (15 April 2018). Berger, Bonnie. ed. "Global proteomics profiling improves drug sensitivity prediction: results from a multi-omics, pan-cancer modeling approach" (in en). Bioinformatics 34 (8): 1353–1362. doi:10.1093/bioinformatics/btx766. ISSN 1367-4803. PMC PMC5905617. PMID 29186355. https://academic.oup.com/bioinformatics/article/34/8/1353/4665420.
- ↑ Zhang, Yaping; He, Chengyan; Qiu, Ling; Wang, Yanmin; Qin, Xuzhen; Liu, Yujie; Li, Zhili (2016). "Serum Unsaturated Free Fatty Acids: A Potential Biomarker Panel for Early-Stage Detection of Colorectal Cancer" (in en). Journal of Cancer 7 (4): 477–483. doi:10.7150/jca.13870. ISSN 1837-9664. PMC PMC4749369. PMID 26918062. http://www.jcancer.org/v07p0477.htm.
- ↑ Giskeødegård, Guro F.; Bertilsson, Helena; Selnæs, Kirsten M.; Wright, Alan J.; Bathen, Tone F.; Viset, Trond; Halgunset, Jostein; Angelsen, Anders et al. (23 April 2013). Monleon, Daniel. ed. "Spermine and Citrate as Metabolic Biomarkers for Assessing Prostate Cancer Aggressiveness" (in en). PLoS ONE 8 (4): e62375. doi:10.1371/journal.pone.0062375. ISSN 1932-6203. PMC PMC3633894. PMID 23626811. https://dx.plos.org/10.1371/journal.pone.0062375.
- ↑ 72.0 72.1 Rajagopala, Seesandra V.; Vashee, Sanjay; Oldfield, Lauren M.; Suzuki, Yo; Venter, J. Craig; Telenti, Amalio; Nelson, Karen E. (1 April 2017). "The Human Microbiome and Cancer" (in en). Cancer Prevention Research 10 (4): 226–234. doi:10.1158/1940-6207.CAPR-16-0249. ISSN 1940-6207. https://aacrjournals.org/cancerpreventionresearch/article/10/4/226/46620/The-Human-Microbiome-and-CancerThe-Human.
- ↑ Helmink, Beth A.; Khan, M. A. Wadud; Hermann, Amanda; Gopalakrishnan, Vancheswaran; Wargo, Jennifer A. (1 March 2019). "The microbiome, cancer, and cancer therapy" (in en). Nature Medicine 25 (3): 377–388. doi:10.1038/s41591-019-0377-7. ISSN 1078-8956. http://www.nature.com/articles/s41591-019-0377-7.
- ↑ Nicora, Giovanna; Vitali, Francesca; Dagliati, Arianna; Geifman, Nophar; Bellazzi, Riccardo (30 June 2020). "Integrated Multi-Omics Analyses in Oncology: A Review of Machine Learning Methods and Tools". Frontiers in Oncology 10: 1030. doi:10.3389/fonc.2020.01030. ISSN 2234-943X. PMC PMC7338582. PMID 32695678. https://www.frontiersin.org/article/10.3389/fonc.2020.01030/full.
- ↑ Chaudhary, Kumardeep; Poirion, Olivier B.; Lu, Liangqun; Garmire, Lana X. (15 March 2018). "Deep Learning–Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer" (in en). Clinical Cancer Research 24 (6): 1248–1259. doi:10.1158/1078-0432.CCR-17-0853. ISSN 1078-0432. PMC PMC6050171. PMID 28982688. https://aacrjournals.org/clincancerres/article/24/6/1248/475/Deep-Learning-Based-Multi-Omics-Integration.
- ↑ Zhang, Qi; Lou, Yu; Yang, Jiaqi; Wang, Junli; Feng, Jie; Zhao, Yali; Wang, Lin; Huang, Xing et al. (1 November 2019). "Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas" (in en). Gut 68 (11): 2019–2031. doi:10.1136/gutjnl-2019-318912. ISSN 0017-5749. PMC PMC6839802. PMID 31227589. https://gut.bmj.com/lookup/doi/10.1136/gutjnl-2019-318912.
- ↑ Seal, Dibyendu Bikash; Das, Vivek; Goswami, Saptarsi; De, Rajat K. (1 July 2020). "Estimating gene expression from DNA methylation and copy number variation: A deep learning regression model for multi-omics integration" (in en). Genomics 112 (4): 2833–2841. doi:10.1016/j.ygeno.2020.03.021. https://linkinghub.elsevier.com/retrieve/pii/S0888754319309449.
- ↑ Ouyang, Xiao; Fan, Qingju; Ling, Guang; Shi, Yu; Hu, Fuyan (6 September 2020). "Identification of Diagnostic Biomarkers and Subtypes of Liver Hepatocellular Carcinoma by Multi-Omics Data Analysis" (in en). Genes 11 (9): 1051. doi:10.3390/genes11091051. ISSN 2073-4425. PMC PMC7566011. PMID 32899915. https://www.mdpi.com/2073-4425/11/9/1051.
- ↑ Huang, Guojun; Wang, Cheng; Fu, Xi (1 November 2021). "Bidirectional deep neural networks to integrate RNA and DNA data for predicting outcome for patients with hepatocellular carcinoma" (in en). Future Oncology 17 (33): 4481–4495. doi:10.2217/fon-2021-0659. ISSN 1479-6694. https://www.futuremedicine.com/doi/10.2217/fon-2021-0659.
- ↑ HEPAVAC Consortium; Löffler, Markus W.; Mohr, Christopher; Bichmann, Leon; Freudenmann, Lena Katharina; Walzer, Mathias; Schroeder, Christopher M.; Trautwein, Nico et al. (1 December 2019). "Multi-omics discovery of exome-derived neoantigens in hepatocellular carcinoma" (in en). Genome Medicine 11 (1): 28. doi:10.1186/s13073-019-0636-8. ISSN 1756-994X. PMC PMC6492406. PMID 31039795. https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-019-0636-8.
- ↑ Shen, Minqian; Xu, Mengyang; Zhong, Fanyi; Crist, McKenzie C.; Prior, Anjali B.; Yang, Kundi; Allaire, Danielle M.; Choueiry, Fouad et al. (20 February 2021). "A Multi-Omics Study Revealing the Metabolic Effects of Estrogen in Liver Cancer Cells HepG2" (in en). Cells 10 (2): 455. doi:10.3390/cells10020455. ISSN 2073-4409. PMC PMC7924215. PMID 33672651. https://www.mdpi.com/2073-4409/10/2/455.
- ↑ Locker, Gershon Y.; Hamilton, Stanley; Harris, Jules; Jessup, John M.; Kemeny, Nancy; Macdonald, John S.; Somerfield, Mark R.; Hayes, Daniel F. et al. (20 November 2006). "ASCO 2006 Update of Recommendations for the Use of Tumor Markers in Gastrointestinal Cancer" (in en). Journal of Clinical Oncology 24 (33): 5313–5327. doi:10.1200/JCO.2006.08.2644. ISSN 0732-183X. https://ascopubs.org/doi/10.1200/JCO.2006.08.2644.
- ↑ Henry, N. Lynn; Hayes, Daniel F. (1 April 2012). "Cancer biomarkers" (in en). Molecular Oncology 6 (2): 140–146. doi:10.1016/j.molonc.2012.01.010. PMC PMC5528374. PMID 22356776. http://doi.wiley.com/10.1016/j.molonc.2012.01.010.
- ↑ Nicolini, Andrea; Ferrari, Paola; Duffy, Michael J. (1 October 2018). "Prognostic and predictive biomarkers in breast cancer: Past, present and future" (in en). Seminars in Cancer Biology 52: 56–73. doi:10.1016/j.semcancer.2017.08.010. https://linkinghub.elsevier.com/retrieve/pii/S1044579X17300524.
- ↑ Chin, Suet-Feung; Santonja, Angela; Grzelak, Marta; Ahn, Soomin; Sammut, Stephen-John; Clifford, Harry; Rueda, Oscar M.; Pugh, Michelle et al. (1 June 2018). "Shallow whole genome sequencing for robust copy number profiling of formalin-fixed paraffin-embedded breast cancers" (in en). Experimental and Molecular Pathology 104 (3): 161–169. doi:10.1016/j.yexmp.2018.03.006. PMC PMC5993858. PMID 29608913. https://linkinghub.elsevier.com/retrieve/pii/S0014480017306470.
- ↑ Raman, Lennart; Van der Linden, Malaïka; Van der Eecken, Kim; Vermaelen, Karim; Demedts, Ingel; Surmont, Veerle; Himpe, Ulrike; Dedeurwaerdere, Franceska et al. (1 December 2020). "Shallow whole-genome sequencing of plasma cell-free DNA accurately differentiates small from non-small cell lung carcinoma" (in en). Genome Medicine 12 (1): 35. doi:10.1186/s13073-020-00735-4. ISSN 1756-994X. PMC PMC7175544. PMID 32317009. https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-020-00735-4.
- ↑ Van Roy, Nadine; Van Der Linden, Malaïka; Menten, Björn; Dheedene, Annelies; Vandeputte, Charlotte; Van Dorpe, Jo; Laureys, Geneviève; Renard, Marleen et al. (15 October 2017). "Shallow Whole Genome Sequencing on Circulating Cell-Free DNA Allows Reliable Noninvasive Copy-Number Profiling in Neuroblastoma Patients" (in en). Clinical Cancer Research 23 (20): 6305–6314. doi:10.1158/1078-0432.CCR-17-0675. ISSN 1078-0432. https://aacrjournals.org/clincancerres/article/23/20/6305/80151/Shallow-Whole-Genome-Sequencing-on-Circulating.
- ↑ Wang, Qi; Peng, Wei-Xian; Wang, Lu; Ye, Li (1 March 2019). "Toward multiomics-based next-generation diagnostics for precision medicine" (in en). Personalized Medicine 16 (2): 157–170. doi:10.2217/pme-2018-0085. ISSN 1741-0541. https://www.futuremedicine.com/doi/10.2217/pme-2018-0085.
- ↑ Menyhárt, Otília; Győrffy, Balázs (2021). "Multi-omics approaches in cancer research with applications in tumor subtyping, prognosis, and diagnosis" (in en). Computational and Structural Biotechnology Journal 19: 949–960. doi:10.1016/j.csbj.2021.01.009. PMC PMC7868685. PMID 33613862. https://linkinghub.elsevier.com/retrieve/pii/S2001037021000131.
- ↑ Asagi, Akinori; Ohta, Koji; Nasu, Junichirou; Tanada, Minoru; Nadano, Seijin; Nishimura, Rieko; Teramoto, Norihiro; Yamamoto, Kazuhide et al. (1 January 2013). "Utility of Contrast-Enhanced FDG-PET/CT in the Clinical Management of Pancreatic Cancer: Impact on Diagnosis, Staging, Evaluation of Treatment Response, and Detection of Recurrence" (in en). Pancreas 42 (1): 11–19. doi:10.1097/MPA.0b013e3182550d77. ISSN 0885-3177. https://journals.lww.com/00006676-201301000-00003.
- ↑ Echle, Amelie; Rindtorff, Niklas Timon; Brinker, Titus Josef; Luedde, Tom; Pearson, Alexander Thomas; Kather, Jakob Nikolas (16 February 2021). "Deep learning in cancer pathology: a new generation of clinical biomarkers" (in en). British Journal of Cancer 124 (4): 686–696. doi:10.1038/s41416-020-01122-x. ISSN 0007-0920. PMC PMC7884739. PMID 33204028. http://www.nature.com/articles/s41416-020-01122-x.
- ↑ Kelly, Christopher J.; Karthikesalingam, Alan; Suleyman, Mustafa; Corrado, Greg; King, Dominic (1 December 2019). "Key challenges for delivering clinical impact with artificial intelligence" (in en). BMC Medicine 17 (1): 195. doi:10.1186/s12916-019-1426-2. ISSN 1741-7015. PMC PMC6821018. PMID 31665002. https://bmcmedicine.biomedcentral.com/articles/10.1186/s12916-019-1426-2.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original citation 19 concerning SNOMED CT had a dead URL; a suitable replacement was used for this version.